Raleigh plot
Raleigh plots, or Rayleigh plots (also called circlegrams[1] and closely related to circular histograms,[2] phasor diagrams,[3] and wind roses[4]), are statistical graphics that serve as graphical representations for a Raleigh test that map a mean vector to a circular plot. Raleigh plots have many applications in the field of chronobiology, such as in studying butterfly migration patterns or protein and gene expression, and in other fields such as geology, cognitive psychology, and physics.
History/Origin
Raleigh plots was first introduced by Lord Rayleigh. The concept of Raleigh plots evolved from Raleigh tests, also introduced by Lord Rayleigh in 1880. The Rayleigh test is a popular statistical test used to measure the concentration of data points around a circle, identifying any unimodal bias in the distribution.[5] Rayleigh plots emerged from this analysis as a means to illustrate the nature of the distribution.
General interpretation
In a Raleigh plot, each individual is assigned a unit vector with a corresponding angle. These unit vectors are averaged together in a Raleigh plot into the mean vector. The length of the mean vector is determined by r (or R), the mean resultant length. r is a measure of concentration, ranging in value between 0 and 1. If the individual angles of the unit vectors are tightly clustered, then the r value will be closer to 1 while if they are widely scattered, then r will be closer to zero.[6] The mean vector is positioned in the center of a circle. Dashes along the circumference of this circle denote desired values. Examples include angles from magnetic north (zero degrees) going clockwise (e.g., 90 degrees from magnetic north, or eastward); times of day, which can also be described in zeitgeber time and circadian time; and phase. Dots on the circumference are usually used to indicate individual unit vectors and their respective angle in regard to the values being measured. Raleigh plots can also use more than one mean vector, particularly if one wants to display the mean vector for different tested groups in the study or to compare mean vectors between groups.[7]
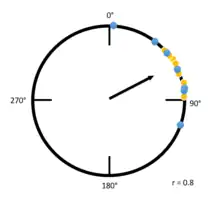
Examples
The example to the left is a Raleigh plot that has a high r value. Blue and yellow dots indicate individuals from different groups being tested, and the position of the dots indicate in which angle from magnetic north each tested individual is traveling. Due to the high overall concentration of individuals going at an angle between zero degrees and ninety degrees, the mean vector is much longer. Compare the figure to the left with the figure below:
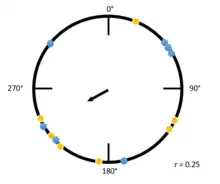
In this second example (to the right), the Raleigh plot has a low r value. Both yellow and blue dots are spread along the circumference of the circle, indicating many individuals are traveling at different angles. The largest cluster of individuals, a group traveling between 180 degrees and 270 degrees, causes the mean vector to be pointed at an angle in that direction. Notably, due to the variability in the direction within this group, the mean vector is much smaller.
Uses in chronobiology studies
Butterfly migration studies
Raleigh plots have been used in chronobiology studies on the biological clocks behind monarch butterfly migration patterns. They are particularly relevant for studying sun compass orientation in migrating butterflies.
In butterfly migration studies, the Raleigh plot maps the orientation of the butterfly when allowed to fly, where the circumference is marked as a compass, with north (N) at the top position. Given the plotted data points, a mean r vector is drawn to indicate the mean orientation of the butterflies in a particular condition.[8][9]
In his studies on the neurobiology of butterfly migration, Steven M. Reppert observes the oriented flight behavior of monarch butterflies. Reppert explains how Raleigh plots are used to handle butterfly orientation data and as tools for the data analysis.[10]
In a 2012 study by Reppert and colleagues on the sufficiency of an antenna for proper time compensation and sun compass orientation in the monarch butterfly, Raleigh plots were used to present the mean flight orientation of butterflies subjected to different study conditions. Along the edge of the circle, degrees 0º (magnetic north) through 360º are shown, the orientation of each butterfly is marked with a dot, and a mean vector is drawn to represent the mean flight orientation recorded.[7] In a 2016 annual review on the neurobiology of monarch butterfly migration, Reppert, Guerra, and Merlin also use Raleigh plots to present butterfly orientation data. The plots were used in the study of the time-compensated sun compass in monarch butterflies.[10] A 2018 review by Reppert and de Roode on the mechanisms of monarch butterfly migration also used Raleigh plots, or circlegrams, to represent butterfly orientation data. Each dot indicates the orientation in which a butterfly individual flew continuously for 5 minutes or longer, and the vector points in the mean direction (some degrees from north) with a magnitude proportional to the mean orientation.[1]
Protein and gene expression studies
Raleigh plots can be used to visualize circadian rhythms in protein or gene expression, and how their phases are affected by other variables or induced conditions.
Jennifer Mohawk, then a postdoc at the University of Texas Southwestern Medical Center, used multiple Raleigh plots to illustrate PER2::LUC expression in her 2013 paper "Methamphetamine and Dopamine Receptor D1 Regulate Entrainment of Murine Circadian Oscillators."[11] Specifically, Mohawk investigated how injections of methamphetamine and D1 antagonist SCH-23390 would shift the peak time of PER2 expression in the liver, lung, pituitary gland, and salivary gland. In these plots, the Raleigh plot can be interpreted as a 24 hour clock with CT 0 at the top of the circle and CT 12 at the bottom of the circle. Each arrow represents the average peak phase of PER2::LUC expression of each group. The strength of the phase clustering is symbolized by the length of the arrow, meaning stronger clustering or closer data points resulting in longer arrows. The individual data points are plotted on the outside of the circle and their unique color and shape resemble the different groups of conditions. At ZT 7 is a pink box that shows the timing of the methamphetamine injection. Mohawk and collaborators compared the angle of the vector, or the mean phase, between the different groups in order to determine if methamphetamine injections induced statistically significant phase changes of PER2::LUC expression within the different glands and organs.[11]
Similarly, Tsedey Mekbib, then a PhD student at the Morehouse School of Medicine, utilized Raleigh plots to depict how the knockdown of SIAH2 impacted the rhythmic expression of all other genes in both males and females. After profiling the entire transcriptome via RNA sequence on the liver at frequent time intervals, the expression of peak timing for all rhythmically expressed genes was plotted on Raleigh plots for each group. These Raleigh plots contain vectors that represent the average peak phase. However, instead of differing the length of the vectors to illustrate the variability in the data points, Mekbib and the other collaborators added a +/- 95% confidence interval that is represented by red range along the circle. In addition to the typical Raleigh plot, the left half of the circle is shaded darker to better visualize the night phase occurring between CT 12 and CT 24.[12]
Suprachiasmatic nucleus (SCN) studies
Raleigh plots can be used to visualize the circadian rhythms in protein or gene expression in the suprachiasmatic nucleus (SCN) specifically, and how they might influence other peripheral tissues.
Raleigh plots were used by Elizabeth Maywood, an English researcher at the MRC Laboratory of Molecular Biology, to visualize how pacemaking activity and synchrony between host SCN cells lacking vasoactive intestinal peptide (VIP) can be restored with a wild-type SCN graft. These plots show vectors that represent the phase of the host SCN cells, measured by PER2::LUC expression. Each plot has a value representing the mean vector length with time points where the cell phases are closer in sync having a value closer to 1. Maywood and collaborators showed that VIP-null host SCN cells synchrony deteriorated over time based on the mean vector length of the Raleigh plots decreasing, and concluded that paracrine signaling from an introduced wild-type SCN graft is sufficient to restore the synchrony between SCN cell pacemakers based on the mean vector length increasing after the graft was introduced.[13]
Mariko Izumo, while working at the University of Texas Southwestern Medical Center, used Raleigh plots to assess the effect of knocking out BMAL1 in the SCN on the circadian expression of PER2, measured using PER2::LUC expression, in the SCN and in peripheral tissues. Izumo and others found that knocking out BMAL1 led to the desynchronization and dampening of PER2 expression in peripheral tissues with Raleigh plots showing different mean phases of the rhythm of PER2 expression. They also show that light/dark cycles and feeding can restore synchrony in peripheral tissues with Raleigh plots showing the same mean phases of the rhythm of PER2 expression.[14]
Similarly, Yongli Shan, also while working at the University of Texas Southwestern Medical Center, used Raleigh plots to show that BMAL1 knockout AVP SCN neurons and VIP SCN neurons show a loss in rhythm in expression of PER2. BMAL1 knockout neurons have data points around the circular diagram and a small mean vector r, while wild-type neurons have a larger mean vector r and data points closer together. Additionally, Shan and others show that intercellular connections to the rest of the SCN was sufficient in restoring rhythmicity in BMAL1 knockout VIP neurons but not AVP neurons with an increase in the mean vector r in VIP neurons but not in AVP neurons.[15]
Uses in other non-chronobiology fields
Raleigh plots or variations on Raleigh plots are used in fields beyond chronobiology. While Raleigh plots visualize a mean vector for data, the variational plots that are closely related to Raleigh plots may visualize histogram data in spokes on a circular chart.
In geology, circular histogram plots or rose diagrams can be used to characterize tectonic plate movements. For example, they may be used to visualize the frequency and direction of fault line motion.[16] In meteorology and climate studies, wind roses are used to present data on the direction, duration, and speed of winds that occurred at a given location. For example, in wind roses released by the Midwestern Regional Climate Center, the length of a spoke in a particular direction, representing a histogram bin, is proportional to the duration of time for which the wind was blowing in that direction, with different colors to show wind speed categories.[4]
Additionally, Raleigh plots can be used in cognitive psychology. Joëlle Provasi, professor at École pratique des hautes études, used them to explain the response of children with or without a lesion in their cerebellum due to surgically removed tumor to a rhythmic stimulus. Provasi and others show a Raleigh plot depicting responses that are close to the stimulus as data points around the top at 0 degrees with a mean vector value close to 1, and a Raleigh plot depicting responses that are irregular with dots spread around the circular plot and a mean vector value closer to 0.[17]
See also
References
- Reppert, Steven M.; de Roode, Jacobus C. (2018-09-10). "Demystifying Monarch Butterfly Migration". Current Biology. 28 (17): R1009–R1022. doi:10.1016/j.cub.2018.02.067. PMID 30205052. S2CID 52186799.
- "circular histogram". Oxford Reference. Retrieved 2023-04-27.
- Rea, Mark S; Bierman, Andrew; Figueiro, Mariana G; Bullough, John D (2008-05-29). "A new approach to understanding the impact of circadian disruption on human health". Journal of Circadian Rhythms. 6: 7. doi:10.1186/1740-3391-6-7. ISSN 1740-3391. PMC 2430544. PMID 18510756.
- "Wind Roses - Charts and Tabular Data | NOAA Climate.gov". www.climate.gov. Retrieved 2023-04-27.
- Landler, Lukas; Ruxton, Graeme D.; Malkemper, E. Pascal (August 2018). "Circular data in biology: advice for effectively implementing statistical procedures". Behavioral Ecology and Sociobiology. 72 (8): 128. doi:10.1007/s00265-018-2538-y. ISSN 0340-5443. PMC 6060829. PMID 30100666.
- Mardia, Kanti V.; Jupp, Peter E. (2000). Directional Statistics. West Sussex, England: John Wiley & Sons Ltd. pp. 1–20. ISBN 0471953334.
- Guerra, Patrick A.; Merlin, Christine; Gegear, Robert J.; Reppert, Steven M. (2012-07-17). "Discordant timing between antennae disrupts sun compass orientation in migratory monarch butterflies". Nature Communications. 3 (1): 958. doi:10.1038/ncomms1965. ISSN 2041-1723. PMC 3962218. PMID 22805565.
- Mouritsen, Henrik; Derbyshire, Rachael; Stalleicken, Julia; Mouritsen, Ole Ø.; Frost, Barrie J.; Norris, D. Ryan (2013-04-30). "An experimental displacement and over 50 years of tag-recoveries show that monarch butterflies are not true navigators". Proceedings of the National Academy of Sciences. 110 (18): 7348–7353. doi:10.1073/pnas.1221701110. ISSN 0027-8424. PMC 3645515. PMID 23569228.
- Franzke, Myriam; Kraus, Christian; Gayler, Maria; Dreyer, David; Pfeiffer, Keram; el Jundi, Basil (2022-02-01). "Stimulus-dependent orientation strategies in monarch butterflies". Journal of Experimental Biology. 225 (3): jeb243687. doi:10.1242/jeb.243687. ISSN 0022-0949. PMC 8918799. PMID 35048981.
- Reppert, Steven M.; Guerra, Patrick A.; Merlin, Christine (2016-03-11). "Neurobiology of Monarch Butterfly Migration". Annual Review of Entomology. 61 (1): 25–42. doi:10.1146/annurev-ento-010814-020855. ISSN 0066-4170. PMID 26473314.
- Mohawk, Jennifer A.; Pezuk, Pinar; Menaker, Michael (2013-04-23). "Methamphetamine and Dopamine Receptor D1 Regulate Entrainment of Murine Circadian Oscillators". PLOS ONE. 8 (4): e62463. doi:10.1371/journal.pone.0062463. ISSN 1932-6203. PMC 3633847. PMID 23626822.
- Mekbib, Tsedey; Suen, Ting-Chung; Rollins-Hairston, Aisha; Smith, Kiandra; Armstrong, Ariel; Gray, Cloe; Owino, Sharon; Baba, Kenkichi; Baggs, Julie E.; Ehlen, J. Christopher; Tosini, Gianluca; DeBruyne, Jason P. (2022-07-05). Liu, Andrew C. (ed.). ""The ubiquitin ligase SIAH2 is a female-specific regulator of circadian rhythms and metabolism"". PLOS Genetics. 18 (7): e1010305. doi:10.1371/journal.pgen.1010305. ISSN 1553-7404. PMC 9286287. PMID 35789210.
- Maywood, Elizabeth S.; Chesham, Johanna E.; O'Brien, John A.; Hastings, Michael H. (2011-08-23). "A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits". Proceedings of the National Academy of Sciences. 108 (34): 14306–14311. doi:10.1073/pnas.1101767108. ISSN 0027-8424. PMC 3161534. PMID 21788520.
- Izumo, Mariko; Pejchal, Martina; Schook, Andrew C; Lange, Ryan P; Walisser, Jacqueline A; Sato, Takashi R; Wang, Xiaozhong; Bradfield, Christopher A; Takahashi, Joseph S (2014-12-19). Ptáček, Louis (ed.). "Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant". eLife. 3: e04617. doi:10.7554/eLife.04617. ISSN 2050-084X. PMC 4298698. PMID 25525750.
- Shan, Yongli; Abel, John H.; Li, Yan; Izumo, Mariko; Cox, Kimberly H.; Jeong, Byeongha; Yoo, Seung-Hee; Olson, David P.; Doyle, Francis J.; Takahashi, Joseph S. (October 2020). "Dual-Color Single-Cell Imaging of the Suprachiasmatic Nucleus Reveals a Circadian Role in Network Synchrony". Neuron. 108 (1): 164–179.e7. doi:10.1016/j.neuron.2020.07.012. PMC 8265161. PMID 32768389.
- Mazur, Stanislaw; Aleksandrowski, Paweł; Gągała, Łukasz; Krzywiec, Piotr; Żaba, Jerzy; Gaidzik, Krzysztof; Sikora, Rafał (June 2020). "Late Palaeozoic strike-slip tectonics versus oroclinal bending at the SW outskirts of Baltica: case of the Variscan belt's eastern end in Poland". International Journal of Earth Sciences. 109 (4): 1133–1160. doi:10.1007/s00531-019-01814-7. ISSN 1437-3254. S2CID 210088268.
- Provasi, Joëlle; Doyère, Valérie; Zélanti, Pierre S.; Kieffer, Virginie; Perdry, Hervé; El Massioui, Nicole; Brown, Bruce L.; Dellatolas, Georges; Grill, Jacques; Droit-Volet, Sylvie (September 2014). "Disrupted sensorimotor synchronization, but intact rhythm discrimination, in children treated for a cerebellar medulloblastoma". Research in Developmental Disabilities. 35 (9): 2053–2068. doi:10.1016/j.ridd.2014.04.024. PMID 24864058.