Replication timing
Replication timing refers to the order in which segments of DNA along the length of a chromosome are duplicated.

DNA replication
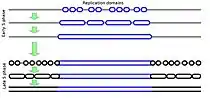
In eukaryotic cells (cells that package their DNA within a nucleus), chromosomes consist of very long linear double-stranded DNA molecules. During the S-phase of each cell cycle (Figure 1), all of the DNA in a cell is duplicated in order to provide one copy to each of the daughter cells after the next cell division. The process of duplicating DNA is called DNA replication, and it takes place by first unwinding the duplex DNA molecule, starting at many locations called DNA replication origins, followed by an unzipping process that unwinds the DNA as it is being copied. However, replication does not start at all the different origins at once. Rather, there is a defined temporal order in which these origins fire. Frequently a few adjacent origins open up to duplicate a segment of a chromosome, followed some time later by another group of origins opening up in an adjacent segment. Replication does not necessarily start at exactly the same origin sites every time, but the segments appear to replicate in the same temporal sequence regardless of exactly where within each segment replication starts. Figure 2 shows a cartoon of how this is generally envisioned to occur, while Figure 3 shows an animation of when different segments replicate in one type of human cell.
Replication timing profiles
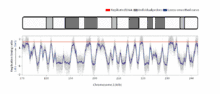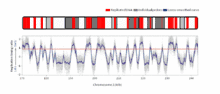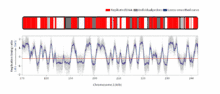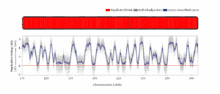
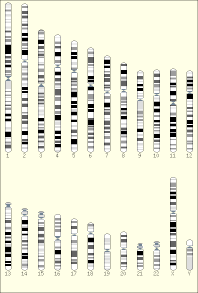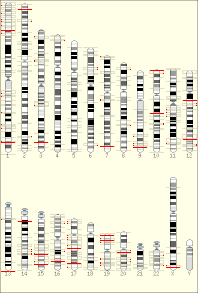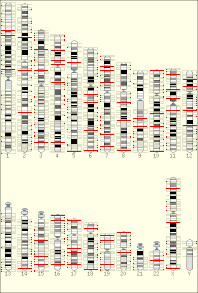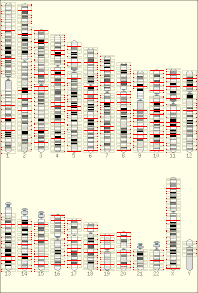
The temporal order of replication of all the segments in the genome, called its replication-timing program, can now be easily measured in two different ways.[1] One way simply measures the amount of the different DNA sequences along the length of the chromosome per cell. Sequences that duplicate first, long before cell division, will be more abundant in each cell than the sequences that replicate last just prior to cell division. The other way is to label newly synthesized DNA with chemically tagged nucleotides that become incorporated into the strands as they are synthesized, and then catch cells at different times during the duplication process and purify the DNA synthesized at each of these times using the chemical tag. In either case, we can measure the amount of the different DNA sequences along the length of the chromosome either directly using a machine that reads how much of each sequence is present or indirectly using a process called microarray hybridization. In any case, the temporal order of replication along the length of each chromosome can be plotted in graphical form to produce a "replication timing profile". Figure 4 shows an example of such a profile across 70,000,000 base pairs of human Chromosome 2.[2]
Replication timing and chromosome structure

At present, very little is known about either the mechanisms orchestrating the timing program or its biological significance. However, it is an intriguing cellular mechanism with links to many poorly understood features of the folding of chromosomes inside the cell nucleus. All eukaryotes have a timing program, and this program is similar in related species.[3][4][5][6][7] This indicates that it is either important itself, or something important influences the program. It is unlikely that replicating DNA in a specific temporal order is necessary simply for the basic purpose of duplicating a DNA molecule. More than likely, it is related to some other chromosomal property or function. Replication timing is correlated with the expression of genes such that the genetic information being utilized in a cell is generally replicated earlier than the information that is not being used. We also know that the replication-timing program changes during development, along with changes in the expression of genes.
For many decades now, it has been known that replication timing is correlated with the structure of chromosomes. For example, female mammals have two X chromosomes. One of these is genetically active, while the other is inactivated early in development. In 1960, J. H. Taylor[8] showed that the active and inactive X chromosomes replicate in a different pattern, with the active X replicating earlier than the inactive X, whereas all the other pairs of chromosomes replicate in the same temporal pattern. It was also noticed by Mary Lyon[9] that the inactive X took on a condensed structure in the nucleus called the Barr body[10] (Figure 5) at the same time during development as the genetic inactivation of the chromosome.
This may not come as too much of a surprise, since the packaging of DNA with proteins and RNA into chromatin takes place immediately after the DNA is synthesized. Therefore, replication timing dictates the time of assembly of chromatin. Less intuitive is the relationship between replication timing and the three-dimensional positioning of chromatin in the nucleus. It is now well-accepted that chromatin is not randomly organized in the cell nucleus, but the positions of each chromosome domain relative to its neighboring domains is characteristic of different cell types, and after this geography is established in each newly formed cell, the chromosome domains do not move appreciably until the next cell division.[11][12] In all multi-cellular organisms where it has been measured, early replication takes place in the interior of the nucleus and the chromatin around the periphery is replicated later. Recently developed methods to measure the points where different parts of chromosomes touch each other are almost perfectly aligned to when they replicate.[3] In other words, regions that are replicated early versus late are packaged in such a way as to be spatially segregated in the nucleus, with the intervening DNA containing regions of reduced origin activity.[7][13] One possibility is that these different compartments within the nucleus, established and maintained without the aid of membranes or physical barriers, set thresholds for the initiation of replication so that the more accessible regions are the first to replicate.[14] Another possibility is that the replication timing of a section of DNA contributes to the packaging of that DNA.[15][16] It has been demonstrated that the protein Rif1 is involved in regulating this process.[17]
Replication timing and disease
Another intriguing aspect of replication timing is that the temporal order of replication is disrupted in most cancers and in many diseases.[18] We do not yet understand the mechanisms behind this link, but it suggests that further research may reveal replication-timing changes as useful biomarkers for such diseases. The fact that it can now be measured with relative ease indicates that we will soon have a wealth of information about where and when large changes in chromosome folding occur during development and in different diseases.
References
- Gilbert DM (2010) Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11: 673-684.
- Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM (2011) Genome-Scale Analysis of Replication Timing: from Bench to Bioinformatics. doi 10.1038/nprot.2011.328.
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, et al. (2010) Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761-770.
- Pope BD, Hiratani I, Gilbert DM (2010) Domain-wide regulation of DNA replication timing during mammalian development. Chromosome Res 18: 127-136.
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, et al. (2009) Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev 23: 589-601.
- Hiratani I, Takebayashi S, Lu J, Gilbert DM (2009) Replication timing and transcriptional control: beyond cause and effect--part II. Curr Opin Genet Dev 19: 142-149.
- Farkash-Amar S, Simon I (2010) Genome-wide analysis of the replication program in mammals. Chromosome Res 18: 115-125.
- Taylor JH (1960) Asynchronous duplication of chromosomes in cultured cells of Chinese hamster. J Biophys Biochem Cytol 7: 455-464.
- Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372-373.
- Barr ML, Bertram EG (1949) A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163: 676.
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 8: 104-115.
- Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T (2003) Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol 160: 685-697.
- Guan Z, Hughes CM, Kosiyatrakul S, Norio P, Sen R, et al. (2009) Decreased replication origin activity in temporal transition regions. J Cell Biol 187: 623-635.
- Gilbert DM (2001) Nuclear position leaves its mark on replication timing. J Cell Biol 152: F11-16.
- Zhang, Jianmin; Xu, Feng; Hashimshony, Tamar; Keshet, Ilana; Cedar, Howard (2002-11-14). "Establishment of transcriptional competence in early and late S phase". Nature. 420 (6912): 198–202. Bibcode:2002Natur.420..198Z. doi:10.1038/nature01150. ISSN 0028-0836. PMID 12432398. S2CID 4359127.
- Lande-Diner, Laura; Zhang, Jianmin; Cedar, Howard (2009-06-26). "Shifts in replication timing actively affect histone acetylation during nucleosome reassembly". Molecular Cell. 34 (6): 767–774. doi:10.1016/j.molcel.2009.05.027. ISSN 1097-2765. PMC 2717743. PMID 19560427.
- Klein, Kyle N.; Zhao, Peiyao A.; Lyu, Xiaowen; Sasaki, Takayo; Bartlett, Daniel A.; Singh, Amar M.; Tasan, Ipek; Zhang, Meng; Watts, Lotte P.; Hiraga, Shin-ichiro; Natsume, Toyoaki; Zhou, Xuemeng; Baslan, Timour; Leung, Danny; Kanemaki, Masato T. (2021-04-23). "Replication timing maintains the global epigenetic state in human cells". Science. 372 (6540): 371–378. Bibcode:2021Sci...372..371K. doi:10.1126/science.aba5545. ISSN 0036-8075. PMC 8173839. PMID 33888635.
- Watanabe Y, Maekawa M (2010) Spatiotemporal regulation of DNA replication in the human genome and its association with genomic instability and disease. Curr Med Chem 17: 222-233.