Rotation distance
In discrete mathematics and theoretical computer science, the rotation distance between two binary trees with the same number of nodes is the minimum number of tree rotations needed to reconfigure one tree into another. Because of a combinatorial equivalence between binary trees and triangulations of convex polygons, rotation distance is equivalent to the flip distance for triangulations of convex polygons.
Rotation distance was first defined by Karel Čulík II and Derick Wood in 1982.[1] Every two n-node binary trees have rotation distance at most 2n − 6, and some pairs of trees have exactly this distance. The computational complexity of computing the rotation distance is unknown.[2]
Definition
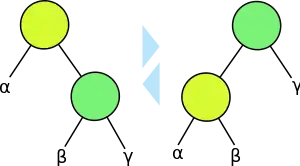
A binary tree is a structure consisting of a set of nodes, one of which is designated as the root node, in which each remaining node is either the left child or right child of some other node, its parent, and in which following the parent links from any node eventually leads to the root node. (In some sources, the nodes described here are called "internal nodes", there exists another set of nodes called "external nodes", each internal node is required to have exactly two children, and each external node is required to have zero children.[1] The version described here can be obtained by removing all the external nodes from such a tree.)
For any node x in the tree, there is a subtree of the same form, rooted at x and consisting of all the nodes that can reach x by following parent links. Each binary tree has a left-to-right ordering of its nodes, its inorder traversal, obtained by recursively traversing the left subtree (the subtree at the left child of the root, if such a child exists), then listing the root itself, and then recursively traversing the right subtree. In a binary search tree, each node is associated with a search key, and the left-to-right ordering is required to be consistent with the order of the keys.[2]
A tree rotation is an operation that changes the structure of a binary tree without changing its left-to-right ordering. Several self-balancing binary search tree data structures use these rotations as a primitive operation in their rebalancing algorithms. A rotation operates on two nodes x and y, where x is the parent of y, and restructures the tree by making y be the parent of x and taking the place of x in the tree. To free up one of the child links of y and make room to link x as a child of y, this operation may also need to move one of the children of y to become a child of x. There are two variations of this operation, a right rotation in which y begins as the left child of x and x ends as the right child of y, and a left rotation in which y begins as the right child of x and x ends as the left child of y.[2]
Any two trees that have the same left-to-right sequence of nodes may be transformed into each other by a sequence of rotations. The rotation distance between the two trees is the number of rotations in the shortest possible sequence of rotations that performs this transformation. It can also be described as the shortest path distance in a rotation graph, a graph that has a vertex for each binary tree on a given left-to-right sequence of nodes and an edge for each rotation between two trees.[2] This rotation graph is exactly the graph of vertices and edges of an associahedron.[3]
Equivalence to flip distance
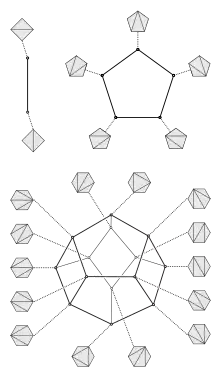
Given a family of triangulations of some geometric object, a flip is an operation that transforms one triangulation to another by removing an edge between two triangles and adding the opposite diagonal to the resulting quadrilateral. The flip distance between two triangulations is the minimum number of flips needed to transform one triangulation into another. It can also be described as the shortest path distance in a flip graph, a graph that has a vertex for each triangulation and an edge for each flip between two triangulations. Flips and flip distances can be defined in this way for several different kinds of triangulations, including triangulations of sets of points in the Euclidean plane, triangulations of polygons, and triangulations of abstract manifolds.
There is a one-to-one correspondence between triangulations of a given convex polygon, with a designated root edge, and binary trees, taking triangulations of n-sided polygons into binary trees with n − 2 nodes. In this correspondence, each triangle of a triangulation corresponds to a node in a binary tree. The root node is the triangle having the designated root edge as one of its sides, and two nodes are linked as parent and child in the tree when the corresponding triangles share a diagonal in the triangulation. Under this correspondence, rotations in binary trees correspond exactly to flips in the corresponding triangulations. Therefore, the rotation distance on (n − 2)-node trees corresponds exactly to flip distance on triangulations of n-sided convex polygons.
Maximum value
Čulík & Wood (1982) define the "right spine" of a binary tree to be the path obtained by starting from the root and following right child links until reaching a node that has no right child. If a tree has the property that not all nodes belong to the right spine, there always exists a right rotation that increases the length of the right spine. For, in this case, there exists at least one node x on the right spine that has a left child y that is not on the right spine. Performing a right rotation on x and y adds y to the right spine without removing any other node from it. By repeatedly increasing the length of the right spine, any n-node tree can be transformed into the unique tree with the same node order in which all nodes belong to the right spine, in at most n − 1 steps. Given any two trees with the same node order, one can transform one into the other by transforming the first tree into a tree with all nodes on the right spine, and then reversing the same transformation of the second tree, in a total of at most 2n − 2 steps. Therefore, as Čulík & Wood (1982) proved, the rotation distance between any two trees is at most 2n − 2.[1]
By considering the problem in terms of flips of convex polygons instead of rotations of trees, Sleator, Tarjan & Thurston (1988) were able to show that the rotation distance is at most 2n − 6. In terms of triangulations of convex polygons, the right spine is the sequence of triangles incident to the right endpoint of the root edge, and the tree in which all vertices lie on the spine corresponds to a fan triangulation for this vertex. The main idea of their improvement is to try flipping both given triangulations to a fan triangulation for any vertex, rather than only the one for the right endpoint of the root edge. It is not possible for all of these choices to simultaneously give the worst-case distance n − 1 from each starting triangulation, giving the improvement.[2]
Sleator, Tarjan & Thurston (1988) also used a geometric argument to show that, for infinitely many values of n, the maximum rotation distance is exactly 2n − 6. They again use the interpretation of the problem in terms of flips of triangulations of convex polygons, and they interpret the starting and ending triangulation as the top and bottom faces of a convex polyhedron with the convex polygon itself interpreted as a Hamiltonian circuit in this polyhedron. Under this interpretation, a sequence of flips from one triangulation to the other can be translated into a collection of tetrahedra that triangulate the given three-dimensional polyhedron. They find a family of polyhedra with the property that (in three-dimensional hyperbolic geometry) the polyhedra have large volume, but all tetrahedra inside them have much smaller volume, implying that many tetrahedra are needed in any triangulation. The binary trees obtained from translating the top and bottom sets of faces of these polyhedra back into trees have high rotation distance, at least 2n − 6.[2]
Subsequently, Pournin (2014) provided a proof that for all n ≥ 11, the maximum rotation distance is exactly 2n − 6. Pournin's proof is combinatorial, and avoids the use of hyperbolic geometry.[3]
Computational complexity
What is the complexity of computing the rotation distance between two trees?
As well as defining rotation distance, Čulík & Wood (1982) asked for the computational complexity of computing the rotation distance between two given trees. The existence of short rotation sequences between any two trees implies that testing whether the rotation distance is at most k belongs to the complexity class NP, but it is not known to be NP-complete, nor is it known to be solvable in polynomial time. Yet, Fordham's algorithm computes rotation distance in linear time, but only allows rotations on the root node and its right child. Fordham's algorithm relies on a classification of nodes into 7 types, and a lookup table is used to find the number of rotations required to transform a node of one type into another.
The rotation distance between any two trees can be lower bounded, in the equivalent view of polygon triangulations, by the number of diagonals that need to be removed from one triangulation and replaced by other diagonals to produce the other triangulation. It can also be upper bounded by twice this number, by partitioning the problem into subproblems along any diagonals shared between both triangulations and then applying the method of Čulík & Wood (1982) to each subproblem. This method provides an approximation algorithm for the problem with an approximation ratio of two.[4] A similar approach of partitioning into subproblems along shared diagonals leads to a fixed-parameter tractable algorithm for computing the rotation distance exactly.[5][6]
Determining the complexity of computing the rotation distance exactly without parameterization remains unsolved, and the best algorithms currently known for the problem run in exponential time.[7] Yet, the existence of Fordham's algorithm strongly suggests a linear time algorithm exists for computing rotation distance.
References
- Čulík, Karel, II; Wood, Derick (1982), "A note on some tree similarity measures", Information Processing Letters, 15 (1): 39–42, doi:10.1016/0020-0190(82)90083-7, MR 0678031
{{citation}}: CS1 maint: multiple names: authors list (link) - Sleator, Daniel D.; Tarjan, Robert E.; Thurston, William P. (1988), "Rotation distance, triangulations, and hyperbolic geometry", Journal of the American Mathematical Society, 1 (3): 647–681, doi:10.1090/S0894-0347-1988-0928904-4, JSTOR 1990951, MR 0928904
- Pournin, Lionel (2014), "The diameter of associahedra", Advances in Mathematics, 259: 13–42, doi:10.1016/j.aim.2014.02.035, MR 3197650
- Cleary, Sean; St. John, Katherine (2010), "A linear-time approximation for rotation distance", Journal of Graph Algorithms and Applications, 14 (2): 385–390, doi:10.7155/jgaa.00212, MR 2740180
- Cleary, Sean; St. John, Katherine (2009), "Rotation distance is fixed-parameter tractable", Information Processing Letters, 109 (16): 918–922, arXiv:0903.0197, doi:10.1016/j.ipl.2009.04.023, MR 2541971, S2CID 125834
- Lucas, Joan M. (2010), "An improved kernel size for rotation distance in binary trees", Information Processing Letters, 110 (12–13): 481–484, doi:10.1016/j.ipl.2010.04.022, MR 2667389
- Kanj, Iyad; Sedgwick, Eric; Xia, Ge (2017), "Computing the flip distance between triangulations", Discrete & Computational Geometry, 58 (2): 313–344, arXiv:1407.1525, doi:10.1007/s00454-017-9867-x, MR 3679938, S2CID 1961246