Rotavirus translation
Rotavirus translation, the process of translating mRNA into proteins, occurs in a different way in Rotaviruses. Unlike the vast majority of cellular proteins in other organisms, in Rotaviruses the proteins are translated from capped but nonpolyadenylated mRNAs. The viral nonstructural protein NSP3 specifically binds the 3'-end consensus sequence of viral mRNAs and interacts with the eukaryotic translation initiation factor eIF4G. The Rotavirus replication cycle occurs entirely in the cytoplasm. Upon virus entry, the viral transcriptase synthesizes capped but nonpolyadenylated mRNA The viral mRNAs bear 5' and 3' untranslated regions (UTR) of variable length and are flanked by two different sequences common to all genes.
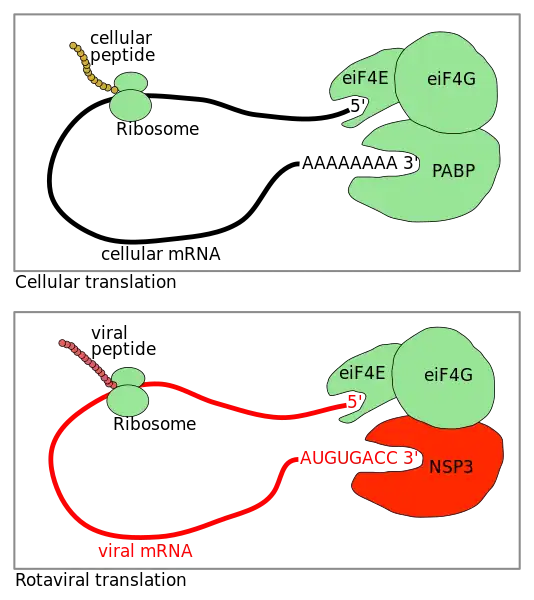
In the group A rotaviruses, the 3'-end consensus sequence UGACC is highly conserved among the 11 genes. Rotavirus NSP3 presents several similarities to PABP; in rotavirus-infected cells, NSP3 can be cross-linked to the 3' end of rotavirus mRNAs and is coimmunoprecipitated with eIF4G. The binding of NSP3A to eIF4G and its specific interaction with the 3' end of viral mRNA brings the viral mRNA and the translation initiation machinery into contact, thus favoring efficient translation of the viral mRNA. NSP3 interacts with the same region of eIF4G as PABP does. As a consequence, during rotavirus infection PABP is evicted from eIF4G, probably impairing the translation of polyadenylated mRNA and leading to the shutoff of cellular mRNA translation observed during rotavirus infection.[1][2]
References
- Poncet, Didier; Carlos Aponte; Jean Cohen (June 1993). "Rotavirus protein NSP3 (NS34) is bound to the 3' end consensus sequence of viral mRNAs in infected cells". J Virol. 67 (6): 3159–3165. doi:10.1128/JVI.67.6.3159-3165.1993. PMC 237654. PMID 8388495.
- Piron, Maria; Patrice Vende; Jean Cohen; Didier Poncet (1998). "Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F". EMBO Journal. 17 (19): 5811–21. doi:10.1093/emboj/17.19.5811. PMC 1170909. PMID 9755181.