S-Adenosylmethionine synthetase enzyme
S-Adenosylmethionine synthetase (EC 2.5.1.6), also known as methionine adenosyltransferase (MAT), is an enzyme that creates S-adenosylmethionine (also known as AdoMet, SAM or SAMe) by reacting methionine (a non-polar amino acid) and ATP (the basic currency of energy).[1]
| Methionine adenosyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 S-adenosylmethionine synthase 2, tetramer, Human | |||||||||
| Identifiers | |||||||||
| EC no. | 2.5.1.6 | ||||||||
| CAS no. | 9012-52-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Function
AdoMet is a methyl donor for transmethylation. It gives away its methyl group and is also the propylamino donor in polyamine biosynthesis. S-adenosylmethionine synthesis can be considered the rate-limiting step of the methionine cycle.[2]
As a methyl donor SAM allows DNA methylation. Once DNA is methylated, it switches the genes off and therefore, S-adenosylmethionine can be considered to control gene expression.[3]
SAM is also involved in gene transcription, cell proliferation, and production of secondary metabolites.[4] Hence SAM synthetase is fast becoming a drug target, in particular for the following diseases: depression, dementia, vacuolar myelopathy, liver injury, migraine, osteoarthritis, and as a potential cancer chemopreventive agent.[5]
This article discusses the protein domains that make up the SAM synthetase enzyme and how these domains contribute to its function. More specifically, this article explores the shared pseudo-3-fold symmetry that makes the domains well-adapted to their functions.[6]
This enzyme catalyses the following chemical reaction
- ATP + L-methionine + H2O phosphate + diphosphate + S-adenosyl-L-methionine
Conserved motifs in the 3'UTR of MAT2A mRNA
A computational comparative analysis of vertebrate genome sequences have identified a cluster of 6 conserved hairpin motifs in the 3'UTR of the MAT2A messenger RNA (mRNA) transcript.[7] The predicted hairpins (named A-F) have strong evolutionary conservation and 3 of the predicted RNA structures (hairpins A, C and D) have been confirmed by in-line probing analysis. No structural changes were observed for any of the hairpins in the presence of metabolites SAM, S-adenosylhomocysteine or L-Methionine. They are proposed to be involved in transcript stability and their functionality is currently under investigation.[7]
Protein overview
The S-adenosylmethionine synthetase enzyme is found in almost every organism bar parasites which obtain AdoMet from their host. Isoenzymes are found in bacteria, budding yeast and even in mammalian mitochondria. Most MATs are homo-oligomers and the majority are tetramers. The monomers are organised into three domains formed by nonconsecutive stretches of the sequence, and the subunits interact through a large flat hydrophobic surface to form the dimers.[8]
S-adenosylmethionine synthetase N terminal domain
| S-adenosylmethionine synthetase N terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
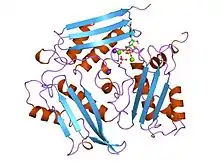 S-adenosylmethionine synthetase with ADP | |||||||||
| Identifiers | |||||||||
| Symbol | S-AdoMet_synt_N | ||||||||
| Pfam | PF00438 | ||||||||
| InterPro | IPR022628 | ||||||||
| PROSITE | PDOC00369 | ||||||||
| SCOP2 | 1mxa / SCOPe / SUPFAM | ||||||||
| |||||||||
In molecular biology the protein domain S-adenosylmethionine synthetase N terminal domain is found at the N-terminal of the enzyme.
N terminal domain function
The N terminal domain is well conserved across different species. This may be due to its important function in substrate and cation binding. The residues involved in methionine binding are found in the N-terminal domain.[8]
N terminal domain structure
The N terminal region contains two alpha helices and four beta strands.[6]
S-adenosylmethionine synthetase Central domain
| S-adenosylmethionine synthetase Central domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 S-adenosylmethionine synthetase with ADP | |||||||||
| Identifiers | |||||||||
| Symbol | S-AdoMet_synt_M | ||||||||
| Pfam | PF02772 | ||||||||
| InterPro | IPR022629 | ||||||||
| PROSITE | PDOC00369 | ||||||||
| SCOP2 | 1mxa / SCOPe / SUPFAM | ||||||||
| |||||||||
Central terminal domain function
The precise function of the central domain has not been fully elucidated, but it is thought to be important in aiding catalysis.
Central domain Structure
The central region contains two alpha helices and four beta strands.[6]
S-adenosylmethionine synthetase, C terminal domain
| S-adenosylmethionine synthetase, C-terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
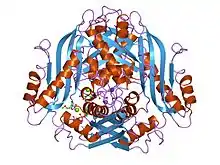 Methionine adenosyltransferase in a complex ADP and l-methionine. | |||||||||
| Identifiers | |||||||||
| Symbol | S-AdoMet_synt_C | ||||||||
| Pfam | PF02773 | ||||||||
| InterPro | IPR022630 | ||||||||
| PROSITE | PDOC00369 | ||||||||
| SCOP2 | 1mxa / SCOPe / SUPFAM | ||||||||
| |||||||||
In molecular biology, the protein domain S-adenosylmethionine synthetase, C-terminal domain refers to the C terminus of the S-adenosylmethionine synthetase
C terminal domain function
The function of the C-terminal domain has been experimentally determined as being important for cytoplasmic localisation. The residues are scattered along the C-terminal domain sequence however once the protein folds, they position themselves closely together.[3]
C terminal domain Structure
The C-terminal domains contains two alpha-helices and four beta-strands.[6]
References
- Horikawa S, Sasuga J, Shimizu K, Ozasa H, Tsukada K (August 1990). "Molecular cloning and nucleotide sequence of cDNA encoding the rat kidney S-adenosylmethionine synthetase". J. Biol. Chem. 265 (23): 13683–6. doi:10.1016/S0021-9258(18)77403-6. PMID 1696256.
- Markham GD, Pajares MA (2009). "Structure-function relationships in methionine adenosyltransferases". Cell Mol Life Sci. 66 (4): 636–48. doi:10.1007/s00018-008-8516-1. PMC 2643306. PMID 18953685.
- Reytor E, Pérez-Miguelsanz J, Alvarez L, Pérez-Sala D, Pajares MA (2009). "Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution". FASEB J. 23 (10): 3347–60. doi:10.1096/fj.09-130187. hdl:10261/55151. PMID 19497982. S2CID 25548921.
- Yoon S, Lee W, Kim M, Kim TD, Ryu Y (2012). "Structural and functional characterization of S-adenosylmethionine (SAM) synthetase from Pichia ciferrii". Bioprocess Biosyst Eng. 35 (1–2): 173–81. doi:10.1007/s00449-011-0640-x. PMID 21989639. S2CID 40318843.
- Kamarthapu V, Rao KV, Srinivas PN, Reddy GB, Reddy VD (2008). "Structural and kinetic properties of Bacillus subtilis S-adenosylmethionine synthetase expressed in Escherichia coli". Biochim Biophys Acta. 1784 (12): 1949–58. doi:10.1016/j.bbapap.2008.06.006. PMID 18634909.
- Takusagawa F, Kamitori S, Misaki S, Markham GD (1996). "Crystal structure of S-adenosylmethionine synthetase". J Biol Chem. 271 (1): 136–47. doi:10.1074/jbc.271.1.136. PMID 8550549.
- Parker BJ, Moltke I, Roth A, Washietl S, Wen J, Kellis M, Breaker R, Pedersen JS (November 2011). "New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes". Genome Res. 21 (11): 1929–43. doi:10.1101/gr.112516.110. PMC 3205577. PMID 21994249.
- Garrido F, Estrela S, Alves C, Sánchez-Pérez GF, Sillero A, Pajares MA (2011). "Refolding and characterization of methionine adenosyltransferase from Euglena gracilis". Protein Expr Purif. 79 (1): 128–36. doi:10.1016/j.pep.2011.05.004. hdl:10261/55441. PMID 21605677.
External links
- Methionine+adenosyltransferase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- EC 2.5.1.6