Single-molecule magnetic sequencing
Magnetic sequencing is a single-molecule sequencing method in development. A DNA hairpin, containing the sequence of interest, is bound between a magnetic bead and a glass surface. A magnetic field is applied to stretch the hairpin open into single strands, and the hairpin refolds after decreasing of the magnetic field. The hairpin length can be determined by direct imaging of the diffraction rings of the magnetic beads using a simple microscope. The DNA sequences are determined by measuring the changes in the hairpin length following successful hybridization of complementary nucleotides.[1]
Single-molecule sequencing vs. Next-generation sequencing
With the development of various next-generation sequencing platforms, there has been a substantial reduction in costs, and increase in throughput of DNA sequencing. However, the majority of the sequencing technologies rely on PCR-based clonal amplification of the DNA molecule in order to bring the signal to a detectable range.[2] Sequencing of amplified clusters, or bulk sequencing in such a propose a read length-dependent phasing problem. During each cycle, not all of the molecules within the bulk have successful incorporation of an additional nucleotide. With increased sequencing cycle, the signal of the lagging molecules will eventually overwhelm the true signal. The phasing problem is a major limitation for the read lengths of the next-generation sequencing technologies. Therefore, there is an increased interest in developing single-molecule sequencing technologies, where no amplification is required. This not only shortens the preparation time for the sequencing libraries, it also has the potential to achieve much longer read lengths, as the lagging molecules with failed extensions can be ignored or considered separately. Previously known single-molecule sequencing technologies include Nanopore sequencing (Oxford Nanopore),[3] SMRT sequencing (Pacific Biosciences),[4] and Heliscope single molecule sequencing (Helicos Biosciences).[5]
Magnetic detection of oligonucleotides hybridized to the DNA hairpin
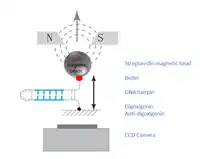
Generation of DNA hairpin
The DNA molecule of interest must be incorporated into a hairpin, and attached to a magnetic bead on one end and to an immobile glass surface on the other end. The hairpin is attached to the glass surface via a digoxigenin-antidigoxigenin bond.[1] The magnetic bead is attached to the opposite end via biotin-streptavidin interaction.[1] Such DNA hairpin setup can be made in two ways:
- 1) In the case of double-stranded DNA molecules (for whole genome sequencing, or targeted sequencing), the DNA fragment is ligated to a DNA loop at one end and a DNA fork structure, labeled with biotin and digoxigenin at the two ends.[1]
- 2) For RNA-seq, the mRNA can be trapped on a poly-T-coated bead, where reverse transcription reaction is performed on the bead to generate a cDNA hairpin.[1]
Measurement of hairpin length
Electromagnets are placed above the sample slide,[6] and an inverted microscope is placed below.[7] The image is captured via a CCD camera and transferred to a computer, where the three-dimensional positions of the magnetic beads are determined. The position of the bead within the horizontal plane of the glass slide, x and y, are determined by real-time correlation of the bead images.[8] The vertical length of the hairpin, measured by the vertical position of the attached magnetic bead, is measured by the bead’s diffraction ring diameter, which increases with distance.[7]
Opening and closing of DNA hairpin
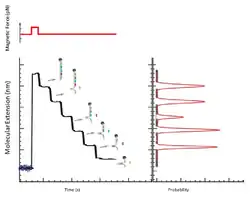
A constant magnetic force is applied to unzip the DNA hairpin, and reducing the force allows the hairpin to rezip.[9] Prior to performing the downstream applications several unzipping and rezipping cycles are performed. While the magnetic force required to unzip and rezip may vary depending on the DNA sequence and hairpin length, their absolute values are not critical as long as they are consistent within a sequencing run.[1]
Detection of hybridization events
When the DNA hairpin is unzipped into single-strand, oligonucleotides complementary to the hairpin sequence are allowed to hybridize. During the time course of the rezipping process, the bound oligonucleotides cause transient blockages.[1] The time course measurement of hairpin length allows for the determination of the exact position of the hybridization, as well as the presence of mismatches between the oligonucleotide and the hairpin.[1]
Applications for hairpin length detection in sequencing
Sequencing by hybridization
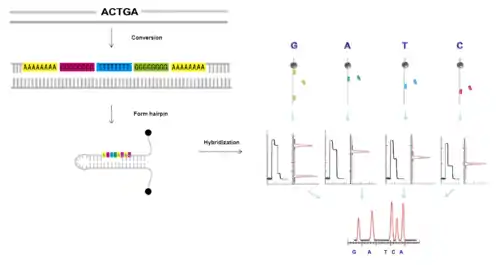
Hybridization is one way to determine the sequence of a DNA strand from detecting the changes in the length of a hairpin. When a probe hybridizes to an open hairpin, complete refolding of the hairpin is stalled, and the position of the hybridized probe can be inferred. Thus the sequence of a DNA fragment of interest can be inferred from overlapping the positions of probes sets, which are allowed to hybridize one by one.[10]
Generation of 8-nt sequence
First, a DNA fragment can be converted into a new sequence in which each original nucleotide is encoded by a specific 8-nt sequence (A8, T8, G8 and C8)[11] and then ligated to a hairpin.
Hybridization of A8, T8, C8, G8 oligonucleotides
After applying a magnetic force, in the unzipped state of the hairpin, a small number of discriminating nucleotides can hybridize to the new individual complementary sequences on the hairpin which can transiently block the refolding of the hairpin.
Map positions
Identification of the blockage positions of the hairpin produced by the hybridization of the discriminating nucleotides can be observed as the pauses in the time course of the hairpin distance measurement. The complete sequence can be reconstructed by the overlapping fragments.
Sequencing by ligation
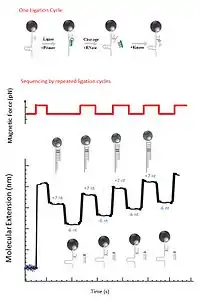
Another application for the magnetic sequencing is using the hairpin end-to-end distance to detect the successive ligation of oligonucleotide.[12] First step of sequencing by ligation is using a primer to extend a DNA fragment. Extension is first attempted with a fragment starting with adenine, which can only be ligated if the next nucleotide on the opposite strand is a thymine. Then fragments starting with cytosine, guanine and thymine are attempted in turn, and the cycle is repeated. The magnetic field is released after each ligation, and then the length of the extended primer is measured. Upon ligation the primer is extended by seven bases, which is resulting in a detectable increase in the hairpin’s end to end distance. RNase cleavage at position 2 is followed by the ligation for the preparation of the next ligation cycle, so that the next ligation is positioned just ahead of the previous one.[10]
7nt primer library
7-nt primer library, 5′-NNNNNNrX-3′, are used in the ligation of a short degenerate oligonucleotide fragmentin, in which N represents any of the four deoxyribonucleotides and Nr represents any of the four ribonucleotides, X is the tested base(A,G,C,T). The ligation to a primer strand of each of the four tested bases in hairpin opening and closing cycles are tested.[1]
Ligation
7-nt primer ligates in the open state of the hairpin, which will block rezipping of the last seven nucleotides and increase the distance between the surface and the magnetic bead by ~5 nm.[1] If the ligation is not successful, no change in the hairpin length is observed.
RNase
RNase cleavage of the last six nucleotides is the next step following the ligation, ultimately extending the primer strand by a single base. Such cleavage allows rezipping of 6 nucleotides of the hairpin, signaled by a decrease in hairpin length of ~4 nm.[1] Therefore, an incorporation of a complementary nucleotide is indicated by an increase in 7 nucleotides (+5 nm) followed by a decrease in 6 nucleotides (-4 nm).[1]
Kinase
After the RNase cleavage of the last six nucleotides, the next step is phosphorylation of the 5'-end via Kinase. Then the next cycle of ligation can be repeated.
Advantages of the magnetic method in single-molecule sequencing
Nature of the detected signal
Many of the competitive single-molecule sequencing methods rely on the incorporation of fluorescently labeled nucleotides.[4][5] In next-generation sequencing, the fluorescence signal of clusters can be easily detected. However, when the same concept is applied to single-molecule sequencing, the largest complication results from the high error rates. Because it is difficult to detect single labeled molecules, these platforms suffer from low signal-to-noise ratios, often resulting in misdetection or non-detection of fluorescent signals.[13] In the case of magnetic sequencing, the signal measured is the changes in distance between two ends of a hairpin. Such signal can be readily detected with standard cameras. Thus, the signals are easier to detect, even without the use of expensive imaging devices.
Relaxation of the experimental constraints for single-nucleotide discrimination
In addition to the nature of the detected signal, other implementations in this platform allows for an even higher signal-to-noise ratio. In the case of magnetic sequencing by hybridization, a set of overlapping tiles is used such that the sequence of each nucleotide is determined by the hybridization of an 8-mer.[1] Therefore, the instrument only requires the sensitivity to detect a change of ~ 6 nm (the length of 8 nucleotides). Similarly, for sequencing by ligation cycles, successful incorporation is characterized by a ~5 nm increase (ligation of a 7-mer) followed by a ~ 4 nm decrease (RNase cleavage of 6-mer) in hairpin length.[1] In this case, the decrease in length in the second step provides additional confirmation for the obtained signal.
Technical considerations
Resolution
With the current methods, the instrumental error in the measured hairpin length is 1-1.5 nm.[1] The length of a basepair, or 2 extended single-stranded nucleotides, is approximately 0.85 nm.[1] Therefore, the resolution of the system is at a few nucleotides. The sources of noise arise from length-dependent Brownian motion of the bead anchored by the extended hairpin, statistical error in bead position determination, and slow mechanical drifts.[1] However, as mentioned earlier, such resolution is sufficient for the current sequencing method because changes in >4 nm are being measured.
Throughput
Through the use of magnetic traps,[6] constant magnetic force can be applied to millions of DNA hairpin-tethered magnetic beads in parallel. The magnetic force can be easily adjusted by changing the distance between the trap and the magnetic beads. The number of eads that can be simultaneously monitored, which determines the read throughput of this platform, is limited by the bead size, length of the tethered DNA hairpin, and the optical resolution limit.[1] Currently, a density of 750 K/mm2 (comparable to an Illumina HiSeq 2000)[14] can be achieved.[1]
Read length
As mentioned above, the noise due to the Brownian fluctuations of the bead increases with length. Robust sequencing tests have yet to be performed to determine the maximum read length of this system. However, the ligation of a 7-mer in the middle of a 1241 nucleotide-long hairpin was successfully detected, suggesting that the current system is sufficient to sequence up to ~500 bp.[1]
Additional limitations
The rate of sequencing or imaging is dependent on the mechanical movement speed of the magnetic beads, which is limited by drag force.[1] Currently, it is possible to measure 10 hairpin open-close cycle per second.[1] Additional complications include the existence of a secondary hairpin structure in the DNA of interest. In such a case the DNA loop to be ligated must be designed such that it its closing is favored over the closing of the endogenous loop in the DNA of interest.[1]
References
- Ding F, Manosas M, Spiering MM, Benkovic SJ, Bensimon D, Allemand JF, Croquette V (March 2012). "Single-molecule mechanical identification and sequencing". Nat. Methods. 9 (4): 367–72. doi:10.1038/nmeth.1925. PMC 3528176. PMID 22406857.
- Shendure J, Ji H (October 2008). "Next-generation DNA sequencing". Nat. Biotechnol. 26 (10): 1135–45. doi:10.1038/nbt1486. PMID 18846087. S2CID 6384349.
- Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, et al. (October 2008). "The potential and challenges of nanopore sequencing". Nat. Biotechnol. 26 (10): 1146–53. doi:10.1038/nbt.1495. PMC 2683588. PMID 18846088.
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. (January 2009). "Real-time DNA sequencing from single polymerase molecules". Science. 323 (5910): 133–8. Bibcode:2009Sci...323..133E. doi:10.1126/science.1162986. PMID 19023044. S2CID 54488479.
- Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, et al. (April 2008). "Single-molecule DNA sequencing of a viral genome". Science. 320 (5872): 106–9. Bibcode:2008Sci...320..106H. doi:10.1126/science.1150427. PMID 18388294. S2CID 16725564.
- Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V (March 1996). "The elasticity of a single supercoiled DNA molecule". Science. 271 (5257): 1835–7. Bibcode:1996Sci...271.1835S. doi:10.1126/science.271.5257.1835. PMID 8596951. S2CID 21790706.
- Gosse C, Croquette V (June 2002). "Magnetic tweezers: micromanipulation and force measurement at the molecular level". Biophys. J. 82 (6): 3314–29. Bibcode:2002BpJ....82.3314G. doi:10.1016/S0006-3495(02)75672-5. PMC 1302119. PMID 12023254.
- Gelles J, Schnapp BJ, Sheetz MP (February 1988). "Tracking kinesin-driven movements with nanometre-scale precision". Nature. 331 (6155): 450–3. Bibcode:1988Natur.331..450G. doi:10.1038/331450a0. PMID 3123999. S2CID 4277258.
- Essevaz-Roulet B, Bockelmann U, Heslot F (October 1997). "Mechanical separation of the complementary strands of DNA". Proc. Natl. Acad. Sci. U.S.A. 94 (22): 11935–40. Bibcode:1997PNAS...9411935E. doi:10.1073/pnas.94.22.11935. PMC 23661. PMID 9342340.
- Linnarsson S (March 2012). "Magnetic sequencing". Nat. Methods. 9 (4): 339–341. doi:10.1038/nmeth.1934. PMID 22453909. S2CID 5328249.
- McNally B, Singer A, Yu Z, Sun Y, Weng Z, Meller A (June 2010). "Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays". Nano Lett. 10 (6): 2237–44. Bibcode:2010NanoL..10.2237M. doi:10.1021/nl1012147. PMC 2883017. PMID 20459065.
- Mir KU, Qi H, Salata O, Scozzafava G (January 2009). "Sequencing by Cyclic Ligation and Cleavage (CycLiC) directly on a microarray captured template". Nucleic Acids Res. 37 (1): e5. doi:10.1093/nar/gkn906. PMC 2615607. PMID 19015154.
- Schadt EE, Turner S, Kasarskis A (October 2010). "A window into third-generation sequencing". Hum. Mol. Genet. 19 (R2): R227–40. doi:10.1093/hmg/ddq416. PMID 20858600.
- Illumina HiSeq 2000 Specifications Archived 2013-03-13 at the Wayback Machine