Virivore
Virivore (equivalently virovore) comes from the English prefix viro- meaning virus, derived from the Latin word for poison, and the suffix -vore from the Latin word vorare, meaning to eat, or to devour;[1] therefore, a virivore is an organism that consumes viruses. Virivory is a well-described process in which organisms, primarily heterotrophic protists,[2][3][4][5][6][7] but also some metazoans[8][9] consume viruses.
Viruses are considered a top predator in marine environments, as they can lyse microbes and release nutrients (i.e. the viral shunt). Viruses also play an important role in the structuring of microbial trophic relationships and regulation of carbon flow.[10][11]
Discovery
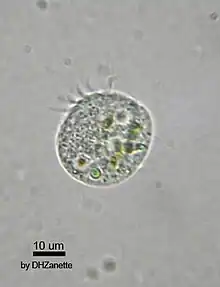
The first described virovore was a small marine flagellate that was shown to ingest and digest virus particles.[2] Subsequently, numerous studies directly and indirectly demonstrated the consumption of virions.[3][4][5][6][7][8][9] In 2022, DeLong et al. showed that over the course of two days the ciliates Halteria and Paramecium reduced chlorovirus plaque-forming units by up to two orders of magnitude, supporting the idea that nutrients were transferred from the viruses to consumers.[7]
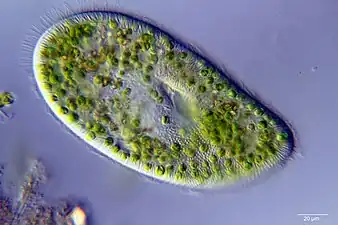
Furthermore, the Halteria population grew with chlorovirus as the only source of nutrition, and grew minimally in the absence of chlorovirus.[7] The Paramecium population, however, did not differ in growth when fed chloroviruses compared to the control group. Since the Paramecium population size remained constant in the presence of only cholorviruses, this indicated that Paramecium is capable of maintaining its population size, but not growing using chlorovirus as the sole carbon source. These data showed that some grazers can grow on viruses, but it does not apply to all grazers. It was estimated that Halteria consumed between 10,000 and 1,000,000 viruses per day.It's known that small protists, such as Halteria and Paramecium, are consumed by zooplankton indicating the movement of viral-derived energy and matter up through the aquatic food web. This contradicts the idea that the viral shunt limits the movement of energy up food webs by cutting off the grazer-microbe interaction. The amount of energy and matter passed up would depend on virion size and nutritional content, which would vary depending on the strain.
Biogeochemical impact

Viruses are the most abundant biological entities in the world's oceans.[12][13][14] The life cycle of a lytic virus is an important process within the worlds oceans for the cycling of dissolved organic matter and particulate organic matter, i.e. the viral shunt.[15][12][13] Viral particles themselves also make up a large proportion of the nitrogen and phosphorus rich particles within the dissolved organic matter pool, as they are made up of lipids, amino acids, nucleic acids, and likely carbon incorporated from host cells.[12][13] It's considered that viruses can complement a grazers diet if ingested, and the microbe is not infected.[14]
General grazing on viruses is widespread throughout the marine environment, with grazing rates as high as 90.3 mL−1 day−1.[12] When both bacteria and viruses are present, viruses can be ingested at rates comparable to bacteria.[2]
Using Oikopleura dioica and Equid alphaherpesvirus 1 (EhV) as a model, scientists estimated the nutritional gain from viruses;[12]
It's suggested that in smaller grazers, viruses could potentially have a more significant impact on host nutrition.[12] For example, in nanoflagellates, the estimated contribution is 9% carbon, 14% nitrogen, and 28% phosphorus.[12]
While smaller bacteria are the ideal food source for grazers due to their size and carbon content, viruses are small, non-motile, and extremely abundant for grazers making them an alternative nutritional choice.[14] For general grazers, to obtain the same amount of carbon from viruses that they get from bacteria, they would need to consume 1000 times more viruses.[14] This does not make viruses the ideal carbon source for grazers. However, there are other benefits to consuming viruses besides growth. Studies show that digested viral particles release amino acids that the grazer can then utilize during their own polypeptide synthesis.[14]
The viral sweep
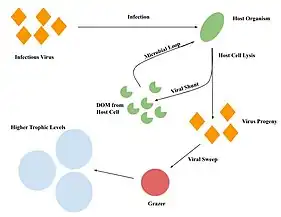
Trophic interactions between grazers, bacteria, and viruses are important in regulating nutrient and organic matter cycling.[14] The viral sweep is a mechanism in which grazers cycle carbon back into the classical food web by ingesting viral particles.[12] Infection of host cells leads to the release of viral progeny, which are subsequently consumed by grazers.[12] Grazers are then consumed by higher trophic organisms, therefore cycling carbon from viruses back into the classical food web and to higher trophic levels.[12]
The viral sweep could be affected by many factors such as the size and abundance of the viral particles.[12] The size of the virus will effect the elemental content of the virus particles.[13] For example, a virus with a larger capsid will contribute more carbon, and viruses with larger genomes will contribute more nitrogen and phosphorus as a result of the increased nucleic acids.[13] Additionally, the impact of the viral sweep could be more significant if grazers preying on bacteria infected with viruses are also considered.[14] Overall, by consuming bacteria and viruses, grazers play an important role in cycling carbon.[14]
Viral grazing
The consumption of viruses is largely based on the feeding behaviour of the organism.
Filter feeding
Filter feeding is a type of suspension feeding.[16] Filter feeders usually actively capture single food particles on cili, hairs, mucus, or other structures.[16] Researchers used Salpingeoca as a model filter feeder to observe change in viral abundance. Salpingeoca produce lorica to help them attach to the substrate.[14] They also have one flagellum to create a water current which transports small particles towards them where tiny pseudopodia engulf the prey particles.[14] When viruses were co-incubated with Salpingeoca, viral abundances decreased steadily over 90 days, showing that filter feeding is an effective mechanism for feeding on viruses.[14]
Grazing on sediment particles
Grazers move over surfaces to gather and ingest food as they go.[16] Researchers used Thaumatomonas coloniensis as a model grazer to observe changes in viral abundances.[14] T. coloniensis glides along the substrate and produce filopodia, which are used to engulf particles associated with the substrate.[14] Over the 90 days, viral abundances steadily decreased when co-incubated with T. coloniensis, showing that grazing is an effective mechanism for feeding on viruses.[14]
Raptorial feeding
Raptorial feeding is a form of active feeding, in which the organism seeks out its prey.[14] Researchers used Goniomonas truncata as a model of raptorial feeding.[14] G. truncata is a cryptomonad that has two flagella which are used to swim close to the substrate searching for food, and they have vacuoles to aid in food uptake.[14] In the presence of G. truncata, viral abundances did not significantly decrease over the course of 90 days.[14] However, this does not exclude the possibility that viral particles are taken up, and then released back into the environment.[14] This data shows that raptorial feeding may not be a method of viral grazing, but it may have other ecological implications in terms of viral transmission.
Selective grazing
Grazing on viruses differs between viruses, and therefore it is subject to selective feeding. Flagellates are capable of ingesting many viruses of different sizes, with the smallest viruses having the lowest ingestion rate.[2] There is huge diversity amongst marine viruses, including size, shape, morphology, and surface charge that may influence the selection, and therefore ingestion rates.[2] Additionally, digestion rates of different viruses by the same flagellate were variable. This implies selection when grazing on viruses.[2] For example, significant differences in virus removal by Tetrahymena pyriformis was observed when the protist was co-incubated with 13 different types of viruses.[17] Additionally, the removal rates for the specific viruses were maintained when the protist was co-incubated with multiple viruses at once.[17] T. pyriformis were able to identify viruses as food, which drives their movement and consumption of certain viruses over others, supporting the idea that some protists are capable of selective grazing.[17]
Impact of viral infection on grazing
Viruses have the capacity to influence the grazing of their host cells during infection, showing that viral infection plays a role in selective grazing.[18][19]
Copepods are a key link in marine food webs as they connect primary and secondary production with higher trophic levels.[18] When phytoplankton Emiliania huxleyi were infected with the coccolithovirus EhV-86, ingestion of the infected cells by the calanoid copepod Acartia tonsa was significantly reduced compared to non-infected cells, indicating selective grazing against infected cells.[18] These results suggest that viral infections reduce grazing, and may potentially reduce food web efficiency by keeping the carbon within the viral shunt-microbial loop, and inhibiting the movement of carbon to higher trophic levels.[18] This emphasizes the importance of the viral sweep for cycling carbon into higher trophic levels.
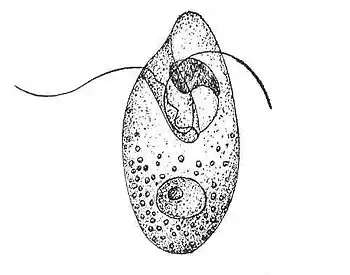
Conversely, Oxyrrhis marina had a grazing preference for virally infected Emiliania huxleyi.[19] It's suggested that the preference of infected cells over non-infected cells is due to physiological changes or change in size of the host cell.[19] O. marina prefer to graze on larger cells as they could potentially get a greater nutritional value from them compared to a smaller cell, which would require the same amount of energy to consume.[19] Infected E. huxleyi exhibit increased cell size compared to non-infected, making them an ideal prey for O. marina.[19] Infected E. huxleyi may also be selected for their palatability as a result of physiological changes during infection.[19] For example, infected cells will have higher nucleic acid content compared to non-infected cells which could improve the nutritional gain to the grazers.[19] Additionally, grazing activity of O. marina has been linked to prey with lower dimethylsulfoniopropionate lyase (DMSP lyase) activity, as they would produce less of the potentially toxic compound acrylate.[19] Virally infected E. huxleyi show reduced levels of DMSP lyase activity, which makes them appealing to O. marina by reducing their exposure to harmful compounds.[19] Lastly, chemical cues such as the release of dimethyl sulfide and hydrogen peroxide during infection likely generate a gradient, making it easier for O. marina to locate the infected E. huxleyi.[19] Preferential grazing on infected cells would make the carbon available to higher trophic levels by sequestering it in particulate form.[19]
Overall, grazing on virus particles and virally infected cells are subject to selective grazing.
Ecological significance
Studies have shown that viruses may be ingested and digested, or ingested and released back into the environment by grazers.[14][20] The observation that grazers could potentially release viruses back into the environment after ingestion could have significant ecological impacts.[20]
Mode of transmission
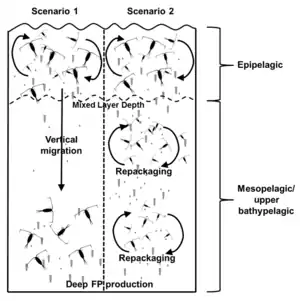
The ingestion and release of viruses could mediate the transmission and dispersal of viruses in the marine environment.[20] Using copepods as the model transmission vector, and EhV as the model virus, Frada et al. identified a potential mechanism of viral dispersal in marine environments.[20]
EhV particles can be consumed by copepods either as individual virion particles or via host cell infection (in this case, infected Emiliania huxleyi).[20] When infected E. huxleyi was co-incubated with copepods, the fecal pellets produced by the copepods contained an average of 4500 EhVs per pellet.[20] These virion containing pellets were then co-incubated with a fresh culture of E. huxleyi, and rapid viral-mediated lysis of the host cells was observed.[20] When EhV particles alone were co-incubated with copepods, i.e. no E. huxleyi, the fecal particles collected did not contain any virion particles.[20] However, when they fed copepods EhV and Thalassiosira weissflogii, a diatom outside the host range of EhV, the fecal pellets collected contained 200 EhVs per pellet.[20] These pellets when co-incubated with a fresh E. huxleyi culture were highly infectious and completely killed the culture.[20] The absence of virion particles in the fecal pellets produced from sole EhV incubation supports the idea that grazers exhibit selective grazing for viruses. EhV can still be taken up by copepods through host cell infection and when in the presence of an ideal food source.[20] Since viral abundance follows bacterial abundance, it is unlikely that there will be a marine environment where viruses will be the sole nutrient source for grazers.[21]
The results of this experiment have significant ecological impacts. Copepods are capable of moving up and down the water column, and migrating short distances between feeding zones.[20][22] Specifically, for copepods and EhV, the movement of copepods can transport viruses into new and non-infected populations of E. huxleyi, promoting bloom demise.[20] Additionally, fecal pellets can sink from the mixed layer into deeper parts of the ocean, where they can be assimilated multiple times.[22] These two scenarios represent potential mechanisms in which viruses can be introduced into new marine environments.
Non-host organisms

Grazers are not the only organisms capable of removing viruses from the water column. Non-host organisms such as anemones, polychaeta larvae, sea squirts, crabs, cockles, oysters, and sponges are all capable of significantly reducing the viral abundance.[23] Sponges were found to have the greatest potential for removing viruses.[23]
The method in which non-host organisms disrupt the viral-host contact is known as transmission interference.[23] Non-host organisms can either have a direct impact by removing the host-organisms, or an indirect one by removing the viruses.[23] These mechanisms cause a reduction in the virus-host contact rates which could significantly impact local microbial population dynamics.[23]
Non-host organisms are capable of removing viruses at rates comparable to natural food particles, bacterial cells, and algal cells, which is higher when compared to grazers that have a viral clearance rate around 4%.[2][23] In regions of high sponge densities, such as coastal and tropical regions, it is likely that the virus removal rate has been underestimated.[23] The effective removal of viruses likely has global ecological impacts that have gone unrecognized.[23]
References
- Government of Canada, Public Services and Procurement Canada (2019-12-06). "-vore – Writing Tips Plus – Writing Tools – Resources of the Language Portal of Canada – Canada.ca". www.noslangues-ourlanguages.gc.ca. Retrieved 2023-04-13.
- González, Juan M.; Suttle, Curtis A. (1993). "Grazing by marine nanoflagellates on viruses and virus-sized particles: Ingestion and digestion" (PDF). Marine Ecology Progress Series. 94 (1): 1–10. Bibcode:1993MEPS...94....1G. doi:10.3354/meps094001.
- Gowing, Marcia M (1993). "Large virus-like particles from vacuoles of phaeodarian radiolarians and from other marine samples" (PDF). Marine Ecology Progress Series. 101: 33–42. Bibcode:1993MEPS..101...33G. doi:10.3354/meps101033.
- Deng, Li; Krauss, Steffen; Feichtmayer, Judith; Hofmann, Roland; Arndt, Hartmut; Griebler, Christian (2014). "Grazing of heterotrophic flagellates on viruses is driven by feeding behaviour". Environmental Microbiology Reports. 6 (4): 325–330. doi:10.1111/1758-2229.12119. PMID 24992530.
- Brown, Julia M.; Labonté, Jessica M.; Brown, Joseph; Record, Nicholas R.; Poulton, Nicole J.; Sieracki, Michael E.; Logares, Ramiro; Stepanauskas, Ramunas (2020). "Single Cell Genomics Reveals Viruses Consumed by Marine Protists". Frontiers in Microbiology. 11: 524828. doi:10.3389/fmicb.2020.524828. PMC 7541821. PMID 33072003.
- Olive, Margot; Moerman, Felix; Fernandez-Cassi, Xavier; Altermatt, Florian; Kohn, Tamar (2022). "Removal of Waterborne Viruses by Tetrahymena pyriformis Is Virus-Specific and Coincides with Changes in Protist Swimming Speed". Environmental Science & Technology. 56 (7): 4062–4070. Bibcode:2022EnST...56.4062O. doi:10.1021/acs.est.1c05518. PMC 8988290. PMID 35258957.
- DeLong, John P.; Van Etten, James L.; Al-Ameeli, Zeina; Agarkova, Irina V.; Dunigan, David D. (2023-01-03). "The consumption of viruses returns energy to food chains". Proceedings of the National Academy of Sciences. 120 (1): e2215000120. Bibcode:2023PNAS..12015000D. doi:10.1073/pnas.2215000120. ISSN 0027-8424. PMID 36574690. S2CID 255219850.
- Hadas, Eran; Marie, Dominique; Shpigel, Muki; Ilan, Micha (2006). "Virus predation by sponges is a new nutrient-flow pathway in coral reef food webs". Limnology and Oceanography. 51 (3): 1548–1550. Bibcode:2006LimOc..51.1548H. doi:10.4319/lo.2006.51.3.1548.
- Welsh, Jennifer E.; Steenhuis, Peter; de Moraes, Karlos Ribeiro; van der Meer, Jaap; Thieltges, David W.; Brussaard, Corina P. D. (2020). "Marine virus predation by non-host organisms". Scientific Reports. 10 (1): 5221. Bibcode:2020NatSR..10.5221W. doi:10.1038/s41598-020-61691-y. PMC 7089979. PMID 32251308.
- Fuhrman JA (June 1999). "Marine viruses and their biogeochemical and ecological effects". Nature. 399 (6736): 541–548. doi:10.1038/21119. PMID 10376593. S2CID 1260399.
- Suttle, Curtis A. (2007). "Marine viruses — major players in the global ecosystem". Nature Reviews Microbiology. 5 (10): 801–812. doi:10.1038/nrmicro1750. ISSN 1740-1534.
- Mayers KM, Kuhlisch C, Basso JT, Saltvedt MR, Buchan A, Sandaa RA (February 2023). Prasad VR (ed.). "Grazing on Marine Viruses and Its Biogeochemical Implications". mBio. 14 (1): e0192121. doi:10.1128/mbio.01921-21. PMC 9973340. PMID 36715508.
- Jover LF, Effler TC, Buchan A, Wilhelm SW, Weitz JS (July 2014). "The elemental composition of virus particles: implications for marine biogeochemical cycles". Nature Reviews. Microbiology. 12 (7): 519–528. doi:10.1038/nrmicro3289. PMID 24931044. S2CID 17421986.
- Deng L, Krauss S, Feichtmayer J, Hofmann R, Arndt H, Griebler C (August 2014). "Grazing of heterotrophic flagellates on viruses is driven by feeding behaviour". Environmental Microbiology Reports. 6 (4): 325–330. doi:10.1111/1758-2229.12119. PMID 24992530.
- Wilhelm, Steven W.; Suttle, Curtis A. (1999). "Viruses and Nutrient Cycles in the Sea". BioScience. 49 (10): 781–788. doi:10.2307/1313569. ISSN 1525-3244.
- "Feeding Stategies". depts.washington.edu. Retrieved 2023-03-29.
- Olive M, Moerman F, Fernandez-Cassi X, Altermatt F, Kohn T (April 2022). "Removal of Waterborne Viruses by Tetrahymena pyriformis Is Virus-Specific and Coincides with Changes in Protist Swimming Speed". Environmental Science & Technology. 56 (7): 4062–4070. doi:10.1021/acs.est.1c05518. PMC 8988290. PMID 35258957.
- Vermont AI, Martínez Martínez J, Waller JD, Gilg IC, Leavitt AH, Floge SA, et al. (September 2016). "Virus infection of Emiliania huxleyi deters grazing by the copepod Acartia tonsa". Journal of Plankton Research. 38 (5): 1194–1205. doi:10.1093/plankt/fbw064.
- Evans C, Wilson WH (2008). "Preferential grazing of Oxyrrhis marina on virus infected Emiliania huxleyi". Limnology and Oceanography. 53 (5): 2035–2040. doi:10.4319/lo.2008.53.5.2035. S2CID 16440441.
- Frada MJ, Schatz D, Farstey V, Ossolinski JE, Sabanay H, Ben-Dor S, et al. (November 2014). "Zooplankton may serve as transmission vectors for viruses infecting algal blooms in the ocean". Current Biology. 24 (21): 2592–2597. doi:10.1016/j.cub.2014.09.031. PMID 25438947. S2CID 2682946.
- Hevroni G, Flores-Uribe J, Béjà O, Philosof A (November 2020). "Seasonal and diel patterns of abundance and activity of viruses in the Red Sea". Proceedings of the National Academy of Sciences of the United States of America. 117 (47): 29738–29747. doi:10.1073/pnas.2010783117. PMC 7703586. PMID 33172994.
- Belcher A, Manno C, Ward P, Henson SA, Sanders R, Tarling GA (2017-03-24). "Copepod faecal pellet transfer through the meso- and bathypelagic layers in the Southern Ocean in spring". Biogeosciences. 14 (6): 1511–1525. doi:10.5194/bg-14-1511-2017. ISSN 1726-4170.
- Welsh JE, Steenhuis P, de Moraes KR, van der Meer J, Thieltges DW, Brussaard CP (March 2020). "Marine virus predation by non-host organisms". Scientific Reports. 10 (1): 5221. doi:10.1038/s41598-020-61691-y. PMC 7089979. PMID 32251308.
Further reading
- Criado, Miguel Ángel (12 January 2023). "Virivores, the organism can eat up to a million viruses a day". EL PAÍS English.
- Schrage, Scott (3 January 2023). "Eating viruses can power growth, reproduction of microorganism". Nebraska Today.