Virtual Cell
Virtual Cell (VCell)[1][2][3][4] is an open-source software platform for modeling and simulation of living organisms, primarily cells. It has been designed to be a tool for a wide range of scientists, from experimental cell biologists to theoretical biophysicists.[5]
| Initial release | October 11, 1999 |
|---|---|
| Stable release | 7.4
/ March 2021 |
| Repository | github |
| Written in | Java, C++, Perl |
| Operating system | Windows, macOS, Linux |
| Platform | IA-32, x64 |
| License | MIT license |
| Website | vcell |
Concept
Virtual Cell is an advanced software platform for modeling and simulating reaction kinetics, membrane transport and diffusion in the complex geometries of cells and multicellular tissues. VCell models have a hierarchical tree structure. The trunk level is the "Physiology" consisting of compartments, species and chemical reactions, and reaction rates that are functions of concentrations. Given initial concentrations of species, VCell can calculate how these concentrations change over time. How these numerical simulations are performed, is determined through a number of "Applications", which specify whether simulations will be deterministic or stochastic, and spatial or compartmental; multiple "Applications" can also specify initial concentrations, diffusion coefficients, flow rates and a variety of modeling assumptions. Thus "Applications" can be viewed as computational experiments to test ideas about the physiological system. Each "Application" corresponds to a mathematical description, which is automatically translated into the VCell Math Description Language. Multiple "Simulations", including parameter scans and changes in solver specifications, can be run within each "Application".
Models can range from the simple to the highly complex, and can represent a mixture of experimental data and purely theoretical assumptions.
The Virtual Cell can be used as a distributed application over the Internet or as a standalone application. The graphical user interface allows construction of complex models in biologically relevant terms: compartment dimensions and shape, molecular characteristics, and interaction parameters. VCell converts the biological description into an equivalent mathematical system of differential equations. Users can switch back-and-forth between the schematic biological view and the mathematical view in the common graphical interface. Indeed, if users desire, they can manipulate the mathematical description directly, bypassing the schematic view. VCell allows users a choice of numerical solvers to translate the mathematical description into software code which is executed to perform the simulations. The results can be displayed on-line, or they can be downloaded to the user's computer in a wide variety of export formats. The Virtual Cell license allows free access to all members of the scientific community.[6]
Users may save their models in the VCell DataBase, which is maintained on servers at U. Connecticut. The VCell Database uses an access control system with permissions to allow users to maintain their models private, share them with select collaborators or make them public. The VCell website maintains a searchable list of models that are public and associated with research publications.
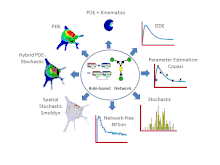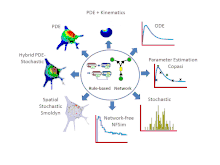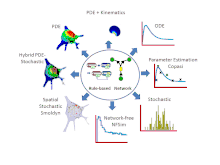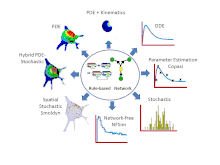
Features
VCell supports the following features:
- Within the "Physiology", models can be specified as reaction networks or reaction rules.[7]
- Simulations can be chosen to either resolve variations of concentrations over space (spatial simulations) or assume concentrations constant across compartments (compartmental simulations).
- For spatial simulations, geometries can be specified by analytic geometry equations, derived from combination of simple shapes or derived from imported images, such as 3D confocal microscope stacks. Utilities for 3D segmentation of image data into regions such as nucleus, mitochondria, cytosol and extracellular are provided.
- Simulations can be based on either integration of differential equations without use of random numbers (deterministic simulations) or be based on random events (stochastic simulations).
- Simulations can be run using a variety of solvers including: 6 ordinary differential equation (ODE) solvers, 2 partial differential equation (PDE) solvers, 4 non-spatial stochastic solvers and Smoldyn[8] for stochastic spatial simulations. VCell also offers a hybrid deterministic/stochastic spatial solver for situations where some species are present in low copy number and others are present in high copy number. Most recently, a network free solver, NFSim, was made available for stochastic simulation of large combinatorially complex rule-based models. Most solvers can be run locally, all solvers can be run remotely on VCell servers.
- For compartmental deterministic models, the best parameter values to fit experimental data can be estimated using algorithms developed by the COPASI software system. These tools are available in VCell.
- Models and simulation setups (so-called Applications) can be stored in local files as Virtual Cell Markup Language (VCML)[9] or stored remotely in the VCell database.
- Models can be imported and exported as Systems Biology Markup Language (SBML)[10]
- Biological pathways can be imported as Biological Pathway Exchange (BioPAX)[11] to build and annotate models.
Biological and related data sources
VCell allows users integrated access to a variety of sources to help build and annotate models:
- Models stored in the VCell database can be made accessible by their authors to some users (shared) or all users (public).
- VCell can import models from the BioModels Database.[12]
- Biological pathways can be imported from Pathway Commons.[13]
- Model elements can be annotated with IDs from Pubmed UniProt (proteins)[14] KEGG (reactions and species) GeneOntology (reactions and species), Reactome (reactions and species) and ChEBI (mostly small molecules).[15]
Development
The Virtual Cell is being developed at the R. D Berlin Center for Cell Analysis and Modeling at the University of Connecticut Health Center.[16] The team is primarily funded through research grants through the National Institutes of Health.
References
- Schaff J, Fink CC, Slepchenko B, Carson JH, Loew LM (September 1997). "A general computational framework for modeling cellular structure and function". Biophysical Journal. 73 (3): 1135–46. Bibcode:1997BpJ....73.1135S. doi:10.1016/S0006-3495(97)78146-3. PMC 1181013. PMID 9284281.
- "Mapping The Mechanisms At The Basis Of Life". Hartford Courant. 23 February 1999. Retrieved 19 March 2012.
- Loew LM, Schaff JC (October 2001). "The Virtual Cell: a software environment for computational cell biology". Trends in Biotechnology. 19 (10): 401–6. doi:10.1016/S0167-7799(01)01740-1. PMID 11587765.
- Cowan AE, Moraru II, Schaff JC, Slepchenko BM, Loew LM (2012). "Spatial modeling of cell signaling networks". Computational Methods in Cell Biology. Vol. 110. Elsevier. pp. 195–221. doi:10.1016/b978-0-12-388403-9.00008-4. ISBN 9780123884039. PMC 3519356. PMID 22482950.
- Moraru II, Schaff JC, Slepchenko BM, Blinov ML, Morgan F, Lakshminarayana A, Gao F, Li Y, Loew LM (September 2008). "Virtual Cell modelling and simulation software environment". IET Systems Biology. 2 (5): 352–62. doi:10.1049/iet-syb:20080102. PMC 2711391. PMID 19045830.
- "VCell - The Virtual Cell". UConn Health Center. Retrieved 22 March 2012.
- Blinov ML, Schaff JC, Vasilescu D, Moraru II, Bloom JE, Loew LM (October 2017). "Compartmental and Spatial Rule-Based Modeling with Virtual Cell". Biophysical Journal. 113 (7): 1365–1372. Bibcode:2017BpJ...113.1365B. doi:10.1016/j.bpj.2017.08.022. PMC 5627391. PMID 28978431.
- "Smoldyn: a spatial stochastic simulator for chemical reaction networks". Retrieved 23 March 2012.
- "VCell Software Architecture - VCML Specification". Retrieved 23 March 2012.
- "Systems Biology Markup Language (SBML)". Retrieved 23 March 2012.
- "BioPAX - Biological Pathway Exchange". Retrieved 23 March 2012.
- "BioModels Database - A Database of Annotated Published Models". Retrieved 23 March 2012.
- "Pathway Commons". Retrieved 23 March 2012.
- "UniProt". Retrieved 23 March 2012.
- "Chemical Entities of Biological Interest (ChEBI)". Retrieved 23 March 2012.
- "The Richard D. Berlin Center for Cell Analysis and Modeling (CCAM)". Retrieved 23 March 2012.