Xeno nucleic acid
Xeno nucleic acids (XNA) are synthetic nucleic acid analogues that have a different sugar backbone than the natural nucleic acids DNA and RNA.[1] As of 2011, at least six types of synthetic sugars have been shown to form nucleic acid backbones that can store and retrieve genetic information. Research is now being done to create synthetic polymerases to transform XNA. The study of its production and application has created a field known as xenobiology.
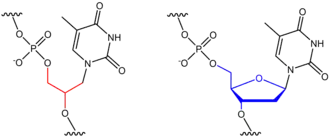
Although the genetic information is still stored in the four canonical base pairs (unlike other nucleic acid analogues), natural DNA polymerases cannot read and duplicate this information. Thus the genetic information stored in XNA is "invisible" and therefore useless to natural DNA-based organisms.[2]
Background
The structure of DNA was discovered in 1953. Around the early 2000s, researchers created a number of exotic DNA-like structures, XNA. XNA is a synthetic polymer that can carry the same information as DNA, but with different molecular constituents. The "X" in XNA stands for "xeno," meaning stranger or alien, indicating the difference in the molecular structure as compared to DNA or RNA.[3]
Not much was done with XNA until the development of special polymerase enzyme, capable of copying XNA from a DNA template as well as copying XNA back into DNA.[3] Pinheiro et al. (2012), for example, has demonstrated such an XNA-capable polymerase that works on sequences of ~100bp in length.[4] More recently, synthetic biologists Philipp Holliger and Alexander Taylor managed to create XNAzymes, the XNA equivalent of a ribozyme, enzymes made of RNA. This demonstrates that XNAs not only store hereditary information, but can also serve as enzymes, raising the possibility that life elsewhere could have begun with something other than RNA or DNA.[5]
Structure
Strands of DNA and RNA are formed by stringing together long chains of molecules called nucleotides. A nucleotide is made up of three chemical components: a phosphate, a five-carbon sugar group (this can be either a deoxyribose sugar—which gives us the "D" in DNA—or a ribose sugar—the "R" in RNA), and one of five standard bases (adenine, guanine, cytosine, thymine or uracil).
The molecules that piece together to form the six xeno nucleic acids are almost identical to those of DNA and RNA, with one exception: in XNA nucleotides, the deoxyribose and ribose sugar groups of DNA and RNA have been replaced with other chemical structures. These substitutions make XNAs functionally and structurally analogous to DNA and RNA despite being unnatural and artificial.
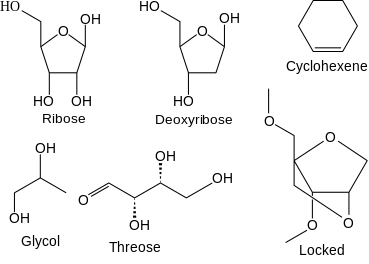
XNA exhibits a variety of structural chemical changes relative to its natural counterparts. Types of synthetic XNA created so far include:[2]
- 1,5-anhydrohexitol nucleic acid (HNA)
- Cyclohexene nucleic acid (CeNA)
- Threose nucleic acid (TNA)
- Glycol nucleic acid (GNA)
- Locked nucleic acid (Locked nucleic acid)
- Peptide nucleic acid (PNA)
- FANA (Fluoro Arabino nucleic acid)
HNA could be used to potentially act as a drug that can recognize and bind to specified sequences. Scientists have been able to isolate HNAs for the possible binding of sequences that target HIV.[6] With cyclohexene nucleic acid, research has shown that CeNAs with stereochemistry similar to the D form can create stable duplexes with itself and RNA. It was shown that CeNAs are not as stable when they form duplexes with DNA.[7]
Implications
The study of XNA is not intended to give scientists a better understanding of biological evolution as it has occurred historically, but rather to explore ways in which we can control and even reprogram the genetic makeup of biological organisms moving forward. XNA has shown significant potential in solving the current issue of genetic pollution in genetically modified organisms.[8] While DNA is incredibly efficient in its ability to store genetic information and lend complex biological diversity, its four-letter genetic alphabet is relatively limited. Using a genetic code of six XNAs rather than the four naturally occurring DNA nucleotide bases yields endless opportunities for genetic modification and expansion of chemical functionality.[9]
The development of various hypotheses and theories about XNAs have altered a key factor in our current understanding of nucleic acids: that heredity and evolution are not limited to DNA and RNA as once thought, but are simply processes that have developed from polymers capable of storing information.[4] Investigations into XNAs will allow for researchers to assess whether DNA and RNA are the most efficient and desirable building blocks of life, or if these two molecules were chosen randomly after evolving from a larger class of chemical ancestors.[10]
Applications
One theory of XNA utilization is its incorporation into medicine as a disease-fighting agent. Some enzymes and antibodies that are currently administered for various disease treatments are broken down too quickly in the stomach or bloodstream. Because XNA is foreign and because it is believed that humans have not yet evolved the enzymes to break them down, XNAs may be able to serve as a more durable counterpart to the DNA and RNA-based treatment methodologies that are currently in use.[11]
Experiments with XNA have already allowed for the replacement and enlargement of this genetic alphabet, and XNAs have shown complementarity with DNA and RNA nucleotides, suggesting potential for its transcription and recombination. One experiment conducted at the University of Florida led to the production of an XNA aptamer by the AEGIS-SELEX (artificially expanded genetic information system - systematic evolution of ligands by exponential enrichment) method, followed by successful binding to a line of breast cancer cells.[12] Furthermore, experiments in the model bacterium E. coli have demonstrated the ability for XNA to serve as a biological template for DNA in vivo.[13]
In moving forward with genetic research on XNAs, various questions must come into consideration regarding biosafety, biosecurity, ethics, and governance/regulation.[2] One of the key questions here is whether XNA in an in vivo setting would intermix with DNA and RNA in its natural environment, thereby rendering scientists unable to control or predict its implications in genetic mutation.[11]
XNA also has potential applications to be used as catalysts, much like RNA has the ability to be used as an enzyme. Researchers have shown XNA is able to cleave and ligate DNA, RNA and other XNA sequences, with the most activity being XNA catalyzed reactions on XNA molecules. This research may be used in determining whether DNA and RNA's role in life emerged through natural selection processes or if it was simply a coincidental occurrence.[14]
XNA may be employed as molecular clamps in quantitative real-time polymerase chain reactions (qPCR) by hybridizing with target DNA sequences.[15] In a study published in PLOS ONE, an XNA-mediated molecular clamping assay detected mutant cell-free DNA (cfDNA) from precancerous colorectal cancer (CRC) lesions and colorectal cancer.[15] XNA may also act as highly specific molecular probes for detection of nucleic acid target sequence.[16]
See also
References
- Schmidt M (2012). Synthetic Biology. John Wiley & Sons. pp. 151–. ISBN 978-3-527-65926-5. Retrieved 9 May 2013.
- Schmidt M (April 2010). "Xenobiology: a new form of life as the ultimate biosafety tool". BioEssays. 32 (4): 322–331. doi:10.1002/bies.200900147. PMC 2909387. PMID 20217844.
- Gonzales R (19 April 2012). "XNA Is Synthetic DNA That's Stronger than the Real Thing". Io9. Retrieved 15 October 2015.
- Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, et al. (April 2012). "Synthetic genetic polymers capable of heredity and evolution". Science. 336 (6079): 341–344. Bibcode:2012Sci...336..341P. doi:10.1126/science.1217622. PMC 3362463. PMID 22517858.
- "World's first artificial enzymes created using synthetic biology". Medical Research Council. 1 December 2014. Archived from the original on 3 January 2018.
- Extance A (19 April 2012). "Polymers perform non-DNA evolution". Royal Society of Chemistry. Retrieved 15 October 2015.
- Gu P, Schepers G, Rozenski J, Van Aerschot A, Herdewijn P (2003). "Base pairing properties of D- and L-cyclohexene nucleic acids (CeNA)". Oligonucleotides. 13 (6): 479–489. doi:10.1089/154545703322860799. PMID 15025914.
- Herdewijn P, Marlière P (June 2009). "Toward safe genetically modified organisms through the chemical diversification of nucleic acids". Chemistry & Biodiversity. 6 (6): 791–808. doi:10.1002/cbdv.200900083. PMID 19554563. S2CID 8572188.
- Pinheiro VB, Holliger P (August 2012). "The XNA world: progress towards replication and evolution of synthetic genetic polymers". Current Opinion in Chemical Biology. 16 (3–4): 245–252. doi:10.1016/j.cbpa.2012.05.198. PMID 22704981.
- Hunter P (May 2013). "XNA marks the spot. What can we learn about the origins of life and the treatment of disease through artificial nucleic acids?". EMBO Reports. 14 (5): 410–413. doi:10.1038/embor.2013.42. PMC 3642382. PMID 23579343.
- "XNA: Synthetic DNA That Can Evolve". Popular Mechanics. 19 April 2012. Retrieved 17 November 2015.
- Sefah K, Yang Z, Bradley KM, Hoshika S, Jiménez E, Zhang L, et al. (January 2014). "In vitro selection with artificial expanded genetic information systems". Proceedings of the National Academy of Sciences of the United States of America. 111 (4): 1449–1454. Bibcode:2014PNAS..111.1449S. doi:10.1073/pnas.1311778111. PMC 3910645. PMID 24379378.
- Pezo V, Liu FW, Abramov M, Froeyen M, Herdewijn P, Marlière P (July 2013). "Binary genetic cassettes for selecting XNA-templated DNA synthesis in vivo". Angewandte Chemie. 52 (31): 8139–8143. doi:10.1002/anie.201303288. PMID 23804524.
- Taylor AI, Pinheiro VB, Smola MJ, Morgunov AS, Peak-Chew S, Cozens C, et al. (February 2015). "Catalysts from synthetic genetic polymers". Nature. 518 (7539): 427–430. Bibcode:2015Natur.518..427T. doi:10.1038/nature13982. PMC 4336857. PMID 25470036.
- Sun Q, Pastor L, Du J, Powell MJ, Zhang A, Bodmer W, et al. (5 October 2021). "A novel xenonucleic acid-mediated molecular clamping technology for early colorectal cancer screening". PLOS ONE. 16 (10): e0244332. Bibcode:2021PLoSO..1644332S. doi:10.1371/journal.pone.0244332. PMC 8491914. PMID 34610014.
- D'Agata R, Giuffrida MC, Spoto G (November 2017). "Peptide Nucleic Acid-Based Biosensors for Cancer Diagnosis". Molecules. 22 (11): 1951. doi:10.3390/molecules22111951. PMC 6150339. PMID 29137122.