Protein ZGRF1
Protein ZGRF1 is a protein encoded in the human by the ZGRF1 gene also known as C4orf21, that has a weight of 236.6 kDa.[5] The ZGRF1 gene product localizes to the cell nucleus and promotes DNA repair by stimulating homologous recombination.[6] This gene shows relatively low expression in most human tissues, with increased expression in situations of chemical dependence. ZGRF1 is orthologous to nearly all eukaryotes. Functional domains of this protein link it to a series of helicases, most notably the AAA_12 and AAA_11 domains.
| ZGRF1 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ZGRF1, C4orf21, zinc finger GRF-type containing 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1918893 HomoloGene: 34708 GeneCards: ZGRF1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Gene
The entire gene is 97,663 base pairs long and has an unprocessed mRNA that is 6,740 nucleotides in length. It consists of 28 exons that encode for a 2104 amino acid protein. 12 splice variants exist for C4orf21.

Locus
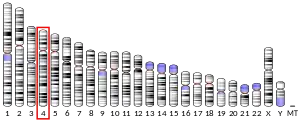
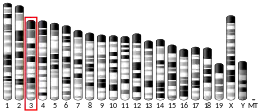
ZGRF1 is located on the fourth chromosome on the 4q25 position near the LARP7 gene. It is encoded for on the minus strand.
Homology and evolution
Homologous domains
ZGRF1 contains a DUF2439 domain (domain of unknown function), zf-GRF domain, and AAA_11 and an AAA_12 domain (ATPases associated with diverse cellular activities). DUF domains are involved in telomere maintenance and meiotic segregation. AAA_11 and AAA_12 contain a P-loop motif which are involved in conjugative transfer proteins. Other helicase domains are also present in c4orf21 orthologs.
Paralogs
There are 9 moderately-related proteins in humans that are paralogous to the ATP-dependent helicase containing domains in the C-terminus of c4orf21 after the 1612th amino acid. A majority of these proteins are in the RNA helicase family. There are no known paralogs to the large N-terminal portion of the protein.
| Paralogous Protein | Protein Name | Amino Acid Identity | Amino Acid Similarity |
|---|---|---|---|
| UPF1 | regulator of nonsense transcripts 1 | 32% | 51% |
| IGHMBP2 | immunoglobulin helicase μ-binding protein 2 | 30% | 47% |
| MOV10 | Moloney Leukemia Virus 10 | 30% | 47% |
| SETX | senataxin | 29% | 43% |
| ZNFX1 | zinc finger, NFX1-type containing 1 | 28% | 47% |
| DNA2 | DNA replication ATP-dependent helicase/nuclease | 26% | 44% |
| PPARG | peroxisome proliferator-activated receptor gamma | 26% | 43% |
| HELZ | helicase with zinc finger domain | 25% | 42% |
| AQR | intron-binding protein Aquarius | 24% | 48% |

Orthologs
Complete orthologs of the c4orf21 gene are found in mammalia. The helicase domain containing C-terminus portion of the gene is conserved across Eukarya.
Protein
Primary sequence
ZGRF1 is 236.6 kDa.

Post-translational modifications
ZGRF1 has experimentally determined phosphorylation sites at the Y38, S137, S140, S325, and S864 positions.
Secondary structure
A weak transmembrane domain is predicted in the TMHMM server with one loop in the C-terminus of the protein prior to the helicase core. This domain contains both ends outside of a membrane.
Tertiary domains and quaternary structure
ZGRF1 has related structures to Upf1, a paralog. These structures have the capability to bind zinc ions and mRNA.

Function and biochemistry
ZGRF1 is a 5’-to-3’ DNA helicase that promotes genome stability by stimulating DNA repair by homologous recombination.[6] Specifically, ZGRF1 facilitates repair of replication-blocking DNA lesions induced by agents such as mitomycin C and camptothecin. Mechanistically, ZGRF1 physically interacts with the RAD51 recombinase and stimulates strand exchange by RAD51-RAD54.
ZGRF1 shares homology in its DUF2439 domain with Saccharomyces cerevisiae Mte1[7][8][9] and Schizosaccharomyces pombe Dbl2,[10][11] which play similar roles in recombinational DNA repair.
The human paralogs to the helicase core of the ZGRF1 gene are associated with translation, transcription, nonsense-mediated mRNA decay, RNA decay, miRNA processing, RISC assembly, and pre-mRNA splicing.[12] These paralogs operate under a SPF1 RNA helicase motif.[13]
Mov10, a paralog, and probable RNA helicase is required for RNA-mediated gene silencing by the RNA-induced silencing complex (RISC). It is also required for both miRNA-mediated translational repression and miRNA-mediated cleavage of complementary mRNAs by RISC, and for RNA-directed transcription and replication of the human hepatitis delta virus (HDV). Mov10 interacts with small capped HDV RNAs derived from genomic hairpin structures that mark the initiation sites of RNA-dependent HDV RNA transcription.
Expression
Expression is relatively low for c4orf21 compared to other proteins. Expression of c4orf21 is slightly elevated compared to its average expression in tissue in the hematopoietic and lymphatic systems, and is above average in the brain also. Lower averages exist in liver, pharynx, and skin tissue.[14]
Transcription factor interactions
The transcriptional start site for ZGRF1 aligns best with ATF, CREB, deltaCREB, E2F, and E2F-1 transcription factor binding sites.
Interacting proteins
C4orf21 shows predicted protein interaction with its AQR, DNA2, IGHMBP2, LOC91431, and SETX paralogs.[15]
Clinical significance
Upon examination of variable GEO profiles, there were many related to Hepatitis and other disorders of the liver. The best correlative studies were those in relation to liver transplant failure.[16][17] ZGRF1 showed significantly increased expression in those who were nicotine dependent versus a control group of non-smokers.[17][18]
A paralog of ZGRF1 was found to inhibit HIV-1 Replication at multiple stages. Mov10 is involved in the biological processes of RNA-mediated gene silencing, transcription, transcription regulation and has hydrolase and helicase activity through ATP and RNA binding.[19]
References
- GRCh38: Ensembl release 89: ENSG00000138658 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000051278 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: Chromosome 4 open reading frame 21".
- Brannvoll A, Xue X, Kwon Y, Kompocholi S, Simonsen AK, Viswalingam KS, et al. (July 2020). "The ZGRF1 Helicase Promotes Recombinational Repair of Replication-Blocking DNA Damage in Human Cells". Cell Reports. 32 (1): 107849. doi:10.1016/j.celrep.2020.107849. PMC 7473174. PMID 32640219.
- Silva S, Altmannova V, Luke-Glaser S, Henriksen P, Gallina I, Yang X, et al. (March 2016). "Mte1 interacts with Mph1 and promotes crossover recombination and telomere maintenance". Genes & Development. 30 (6): 700–17. doi:10.1101/gad.276204.115. PMC 4803055. PMID 26966248.
- Xue X, Papusha A, Choi K, Bonner JN, Kumar S, Niu H, et al. (March 2016). "Differential regulation of the anti-crossover and replication fork regression activities of Mph1 by Mte1". Genes & Development. 30 (6): 687–99. doi:10.1101/gad.276139.115. PMC 4803054. PMID 26966246.
- Yimit A, Kim T, Anand RP, Meister S, Ou J, Haber JE, et al. (May 2016). "MTE1 Functions with MPH1 in Double-Strand Break Repair". Genetics. 203 (1): 147–57. doi:10.1534/genetics.115.185454. PMC 4858770. PMID 26920759.
- Yu Y, Ren JY, Zhang JM, Suo F, Fang XF, Wu F, Du LL (June 2013). "A proteome-wide visual screen identifies fission yeast proteins localizing to DNA double-strand breaks". DNA Repair. 12 (6): 433–43. doi:10.1016/j.dnarep.2013.04.001. PMID 23628481.
- Polakova S, Molnarova L, Hyppa RW, Benko Z, Misova I, Schleiffer A, et al. (June 2016). Lichten M (ed.). "Dbl2 Regulates Rad51 and DNA Joint Molecule Metabolism to Ensure Proper Meiotic Chromosome Segregation". PLOS Genetics. 12 (6): e1006102. doi:10.1371/journal.pgen.1006102. PMC 4909299. PMID 27304859.
- Jankowsky E (Jan 2011). "RNA helicases at work: binding and rearranging". Trends in Biochemical Sciences. 36 (1): 19–29. doi:10.1016/j.tibs.2010.07.008. PMC 3017212. PMID 20813532.
- Fairman-Williams ME, Guenther UP, Jankowsky E (Jun 2010). "SF1 and SF2 helicases: family matters". Current Opinion in Structural Biology. 20 (3): 313–24. doi:10.1016/j.sbi.2010.03.011. PMC 2916977. PMID 20456941.
- "c4orf21". Expression Atlas. Archived from the original on July 6, 2013. Retrieved 16 May 2013.
- Anon. "Predicted protein interactions between paralogs and c4orf21". C4orf21 Gene - GeneCards. Retrieved 16 May 2013.
- Nissim O, Melis M, Diaz G, Kleiner DE, Tice A, Fantola G, Zamboni F, Mishra L, Farci P (2012). "Liver regeneration signature in hepatitis B virus (HBV)-associated acute liver failure identified by gene expression profiling". PLOS ONE. 7 (11): e49611. Bibcode:2012PLoSO...749611N. doi:10.1371/journal.pone.0049611. PMC 3504149. PMID 23185381.
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A (Jan 2013). "NCBI GEO: archive for functional genomics data sets--update". Nucleic Acids Research. 41 (Database issue): D991–5. doi:10.1093/nar/gks1193. PMC 3531084. PMID 23193258.
- Philibert RA, Ryu GY, Yoon JG, Sandhu H, Hollenbeck N, Gunter T, Barkhurst A, Adams W, Madan A (Jul 2007). "Transcriptional profiling of subjects from the Iowa adoption studies". American Journal of Medical Genetics Part B. 144B (5): 683–90. doi:10.1002/ajmg.b.30512. PMID 17342724. S2CID 6002286.
- Burdick R, Smith JL, Chaipan C, Friew Y, Chen J, Venkatachari NJ, Delviks-Frankenberry KA, Hu WS, Pathak VK (Oct 2010). "P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages". Journal of Virology. 84 (19): 10241–53. doi:10.1128/JVI.00585-10. PMC 2937795. PMID 20668078.
External links
- Human ZGRF1 genome location and ZGRF1 gene details page in the UCSC Genome Browser.
Further reading
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Séraphin B, Le Hir H, Andersen GR (Sep 2006). "Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA". Science. 313 (5795): 1968–72. Bibcode:2006Sci...313.1968A. doi:10.1126/science.1131981. PMID 16931718. S2CID 26409491.
- Le Hir H, Andersen GR (Feb 2008). "Structural insights into the exon junction complex". Current Opinion in Structural Biology. 18 (1): 112–9. doi:10.1016/j.sbi.2007.11.002. PMID 18164611.
- Schwer B (Jun 2008). "A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release". Molecular Cell. 30 (6): 743–754. doi:10.1016/j.molcel.2008.05.003. PMC 2465764. PMID 18570877.
- Lohman TM, Tomko EJ, Wu CG (May 2008). "Non-hexameric DNA helicases and translocases: mechanisms and regulation". Nature Reviews Molecular Cell Biology. 9 (5): 391–401. doi:10.1038/nrm2394. PMID 18414490. S2CID 6176756.
- Liu F, Putnam A, Jankowsky E (Dec 2008). "ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding". Proceedings of the National Academy of Sciences of the United States of America. 105 (51): 20209–20214. Bibcode:2008PNAS..10520209L. doi:10.1073/pnas.0811115106. PMC 2629341. PMID 19088201.
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S (Apr 2006). "Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa". Cell. 125 (2): 287–300. doi:10.1016/j.cell.2006.01.054. PMID 16630817. S2CID 14994628.
- Hirano M, Quinzii CM, Mitsumoto H, Hays AP, Roberts JK, Richard P, Rowland LP (May 2011). "Senataxin mutations and amyotrophic lateral sclerosis". Amyotrophic Lateral Sclerosis. 12 (3): 223–7. doi:10.3109/17482968.2010.545952. PMC 7528023. PMID 21190393.
- Wang X, Han Y, Dang Y, Fu W, Zhou T, Ptak RG, Zheng YH (May 2010). "Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication". The Journal of Biological Chemistry. 285 (19): 14346–55. doi:10.1074/jbc.M110.109314. PMC 2863248. PMID 20215113.