ZNF837
ZNF837 is a protein that in humans is encoded by the ZNF837 gene,[3] is located at 19q13.431 with minus strand orientation.[4] ZNF837 protein is characterized as a C2H2-type zinc finger protein.[5]
| ZNF837 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ZNF837, zinc finger protein 837 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | HomoloGene: 138182 GeneCards: ZNF837 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Homology and Evolution
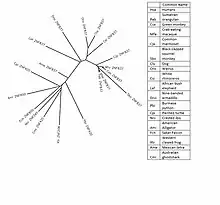
The human ZNF837 has homologs present in many mammals and seen more distantly. All homologs are chordates. All contain both COG5048 and Zf-C2H2_2 domains. The areas that these domains are found contain the highest conservation rates. In humans, 5 Zf-H2C2 double domains[6] and 2 COG5048 domains[7] are present.
The protein sequence is fast evolving among these homologs.
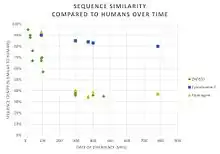
ZNF837 has numerous paralogs in humans, all of which are zinc finger proteins.
Human ZNF837
In humans, there are no other aliases, and its neighboring genes are MIR4754, A1BG, and RPS5. ZNF837 mRNA that is made into function protein contains 1921 nucleotides, of which 222-1817 are translated to a protein containing 3 exons. The protein consists of 531 amino acids[8] with a weight of 58,078 Da with an isoelectric point at 9.525.[9]
Gene Variants
There are 2 transcript variants. Transcript variant 1, 2050 base pairs in length, is non-coding due to a nonsense-mediated mRNA decay .[10] Transcript variant 2 is made into the functional protein due to an alternate splice site[11]
Post Translational
It is predicted via high conservation to have 4 phosphorylation sites[12] at T386, T455, S460, Y503.[13] The internal structure is includes combination of alpha helices, beta sheets and mainly random coils.[14]
Expression
ZNF837 has observed in the pancreas, liver, uterus, and muscle cells. In all cases concentration is low.[15] However, the expression of ZNF837 is seen to have the most impact is when looking at normal vs diseased state. There is a consistent change that is able to be seen.
References
- GRCh38: Ensembl release 89: ENSG00000152475 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Strausberg, RL; Feingold, EA; Grouse, LH; et al. (December 2002). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proceedings of the National Academy of Sciences. 99 (26): 16899–16903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- "ZNF837 zinc finger protein 837 [Homo sapiens (human)] - Gene - NCBI".
- Pieler, T.; Bellefroid, E. (1994). "Perspectives on zinc finger protein function and evolution - an update". Molecular Biology Reports. 20 (1): 1–8. doi:10.1007/bf00999848. PMID 7531280. S2CID 22962225.
- Iuchi, S (2001). "Three classes of C2H2 zinc finger proteins". Cellular and Molecular Life Sciences. 58 (4): 625–635. doi:10.1007/pl00000885. PMID 11361095. S2CID 6522993.
- "NCBI Conserved Domain Search".
- "ZNF837 protein [Homo sapiens] - Protein - NCBI".
- "ZNF837 (human)".
- Maquat, Lynne E (2002). "Nonsense-mediated mRNA decay". Current Biology. 12 (6): R196–R197. doi:10.1016/S0960-9822(02)00747-9. PMID 11909543.
- "ZNF837 (human)".
- Blom, Nikolaj; Gammeltoft, Steen; Brunak, Søren (1999). "Sequence and structure-based prediction of eukaryotic protein phosphorylation sites 1". Journal of Molecular Biology. 294 (5): 1351–1362. doi:10.1006/jmbi.1999.3310. PMID 10600390.
- Guo, Ailan et al. Serine, Threonine, and Tyrosine Phosphorylation Sites. Patent US20110045603 A1. 20 Apr. 2010. Print.
- Kelley, LA; Sternberg, MJE (2009). "Protein structure prediction on the Web: a case study using the Phyre server". Nature Protocols. 4 (3): 363–371. doi:10.1038/nprot.2009.2. hdl:10044/1/18157. PMID 19247286. S2CID 12497300.
- "EST Profile - Hs.222236".