Cas9
Cas9 (CRISPR associated protein 9, formerly called Cas5, Csn1, or Csx12) is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in genetic engineering applications. Its main function is to cut DNA and thereby alter a cell's genome. The CRISPR-Cas9 genome editing technique was a significant contributor to the Nobel Prize in Chemistry in 2020 being awarded to Emmanuelle Charpentier and Jennifer Doudna.[2]
| CRISPR-associated endonuclease Cas9 | |||||||
|---|---|---|---|---|---|---|---|
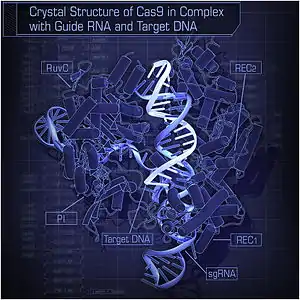 Crystal structure of S. pyogenes Cas9 in complex with sgRNA and its target DNA at 2.5 Å resolution.[1] | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | cas9 | ||||||
| Alt. symbols | SpCas9 | ||||||
| Entrez | 901176 | ||||||
| PDB | 4OO8 | ||||||
| RefSeq (mRNA) | NC_002737.2 | ||||||
| RefSeq (Prot) | NP_269215.1 | ||||||
| UniProt | Q99ZW2 | ||||||
| Other data | |||||||
| EC number | 3.1.-.- | ||||||
| Chromosome | Genomic: 0.85 - 0.86 Mb | ||||||
| |||||||
| Cas9 | |
|---|---|
| Identifiers | |
| Symbol | ? |
| InterPro | IPR028629 |
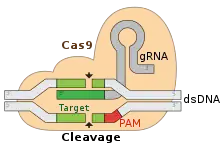
More technically, Cas9 is a dual RNA-guided DNA endonuclease enzyme associated with the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) adaptive immune system in Streptococcus pyogenes.[3][4] S. pyogenes utilizes CRISPR to memorize and Cas9 to later interrogate and cleave foreign DNA, such as invading bacteriophage DNA or plasmid DNA.[4][5][6][7] Cas9 performs this interrogation by unwinding foreign DNA and checking for sites complementary to the 20 nucleotide spacer region of the guide RNA (gRNA). If the DNA substrate is complementary to the guide RNA, Cas9 cleaves the invading DNA. In this sense, the CRISPR-Cas9 mechanism has a number of parallels with the RNA interference (RNAi) mechanism in eukaryotes.
Apart from its original function in bacterial immunity, the Cas9 protein has been heavily utilized as a genome engineering tool to induce site-directed double-strand breaks in DNA. These breaks can lead to gene inactivation or the introduction of heterologous genes through non-homologous end joining and homologous recombination respectively in many laboratory model organisms. Alongside zinc finger nucleases and Transcription activator-like effector nuclease (TALEN) proteins, Cas9 is becoming a prominent tool in the field of genome editing.
Cas9 has gained traction in recent years because it can cleave nearly any sequence complementary to the guide RNA.[4] Because the target specificity of Cas9 stems from the guide RNA:DNA complementarity and not modifications to the protein itself (like TALENs and zinc fingers), engineering Cas9 to target new DNA is straightforward.[8] Versions of Cas9 that bind but do not cleave cognate DNA can be used to locate transcriptional activator or repressors to specific DNA sequences in order to control transcriptional activation and repression.[9][10] Native Cas9 requires a guide RNA composed of two disparate RNAs that associate – the CRISPR RNA (crRNA), and the trans-activating crRNA (tracrRNA).[3] Cas9 targeting has been simplified through the engineering of a chimeric single guide RNA (chiRNA). Scientists have suggested that Cas9-based gene drives may be capable of editing the genomes of entire populations of organisms.[11] In 2015, Cas9 was used to modify the genome of human embryos for the first time.[12]
CRISPR-mediated immunity
To survive in a variety of challenging, inhospitable habitats that are filled with bacteriophages, bacteria and archaea have evolved methods to evade and fend off predatory viruses. This includes the CRISPR system of adaptive immunity. In practice, CRISPR/Cas systems act as self-programmable restriction enzymes. CRISPR loci are composed of short, palindromic repeats that occur at regular intervals composed of alternate CRISPR repeats and variable CRISPR spacers between 24 and 48 nucleotides long. These CRISPR loci are usually accompanied by adjacent CRISPR-associated (cas) genes. In 2005, it was discovered by three separate groups that the spacer regions were homologous to foreign DNA elements, including plasmids and viruses. These reports provided the first biological evidence that CRISPRs might function as an immune system.
Cas9 has been used often as a genome-editing tool. Cas9 has been used in recent developments in preventing viruses from manipulating hosts' DNA. Since the CRISPR-Cas9 was developed from bacterial genome systems, it can be used to target the genetic material in viruses. The use of the enzyme Cas9 can be a solution to many viral infections. Cas9 possesses the ability to target specific viruses by the targeting of specific strands of the viral genetic information. More specifically the Cas9 enzyme targets certain sections of the viral genome that prevents the virus from carrying out its normal function.[13] Cas9 has also been used to disrupt the detrimental strand of DNA and RNA that cause diseases and mutated strands of DNA. Cas9 has already showed promise in disrupting the effects of HIV-1. Cas9 has been shown to suppress the expression of the long terminal repeats in HIV-1. When introduced into the HIV-1 genome Cas9 has shown the ability to mutate strands of HIV-1.[14][15] Cas9 has also been used in the treatment of Hepatitis B through targeting of the ends of certain of long terminal repeats in the Hepatitis B viral genome.[16] Cas9 has been used to repair the mutations causing cataracts in mice.
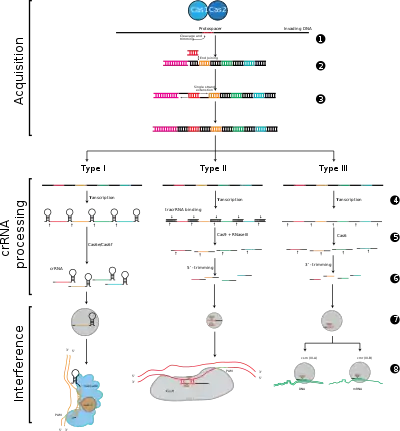
CRISPR-Cas systems are divided into three major types (type I, type II, and type III) and twelve subtypes, which are based on their genetic content and structural differences. However, the core defining features of all CRISPR-Cas systems are the cas genes and their proteins: cas1 and cas2 are universal across types and subtypes, while cas3, cas9, and cas10 are signature genes for type I, type II, and type III, respectively.
Adaptation
Adaptation involves recognition and integration of spacers between two adjacent repeats in the CRISPR locus. The "Protospacer" refers to the sequence on the viral genome that corresponds to the spacer. A short stretch of conserved nucleotides exists proximal to the protospacer, which is called the protospacer adjacent motif (PAM). The PAM is a recognition motif that is used to acquire the DNA fragment.[7] In type II, Cas9 recognizes the PAM during adaptation in order to ensure the acquisition of functional spacers.[5]
Loss of spacers and even groups of several have also been observed by Aranaz et al. 2004 and Pourcel et al. 2007. This probably occurs through homologous recombination of the between-repeat material.[17]
CRISPR processing/biogenesis
CRISPR expression includes the transcription of a primary transcript called a CRISPR RNA (pre-crRNA), which is transcribed from the CRISPR locus by RNA polymerase. Specific endoribonucleases then cleave the pre-crRNAs into small CRISPR RNAs (crRNAs).[18]
Interference/immunity
Interference involves the crRNAs within a multi-protein complex called CASCADE, which can recognize and specifically base-pair with regions of inserting complementary foreign DNA. The crRNA-foreign nucleic acid complex is then cleaved, however if there are mismatches between the spacer and the target DNA, or if there are mutations in the PAM, then cleavage will not be initiated. In the latter scenario, the foreign DNA is not targeted for attack by the cell, thus the replication of the virus proceeds and the host is not immune to viral infection. The interference stage can be mechanistically and temporally distinct from CRISPR acquisition and expression, yet for complete function as a defense system, all three phases must be functional.[19]
Stage 1: CRISPR spacer integration. Protospacers and protospacer-associated motifs (shown in red) are acquired at the "leader" end of a CRISPR array in the host DNA. The CRISPR array is composed of spacer sequences (shown in colored boxes) flanked by repeats (black diamonds). This process requires Cas1 and Cas2 (and Cas9 in type II[5]), which are encoded in the cas locus, which are usually located near the CRISPR array.
Stage 2: CRISPR expression. Pre-crRNA is transcribed starting at the leader region by the host RNA polymerase and then cleaved by Cas proteins into smaller crRNAs containing a single spacer and a partial repeat (shown as hairpin structure with colored spacers).
Stage 3: CRISPR interference. crRNA with a spacer that has strong complementarity to the incoming foreign DNA begins a cleavage event (depicted with scissors), which requires Cas proteins. DNA cleavage interferes with viral replication and provides immunity to the host. The interference stage can be functionally and temporarily distinct from CRISPR acquisition and expression (depicted by white line dividing the cell).
Transcription deactivation using dCas9
dCas9, also referred to as endonuclease deficient Cas9 can be utilized to edit gene expression when applied to the transcription binding site of the desired section of a gene. The optimal function of dCas9 is attributed to its mode of action. Gene expression is inhibited when nucleotides are no longer added to the RNA chain and therefore terminating elongation of that chain, and as a result affects the transcription process. This process occurs when dCas9 is mass-produced so it is able to affect the most genes at any given time via a sequence specific guide RNA molecule. Since dCas9 appears to down regulate gene expression, this action is amplified even more when it is used in conjunction with repressive chromatin modifier domains.[20] The dCas9 protein has other functions outside of the regulation of gene expression. A promoter can be added to the dCas9 protein which allows them to work with each other to become efficient at beginning or stopping transcription at different sequences along a strand of DNA. These two proteins are specific in where they act on a gene. This is prevalent in certain types of prokaryotes when a promoter and dCas9 align themselves together to impede the ability of elongation of polymer of nucleotides coming together to form a transcribed piece of DNA. Without the promoter, the dCas9 protein does not have the same effect by itself or with a gene body.[21]
When examining the effects of repression of transcription further, H3K27, an amino acid component of a histone, becomes methylated through the interaction of dCas9 and a peptide called FOG1. Essentially, this interaction causes gene repression on the C + N terminal section of the amino acid complex at the specific junction of the gene, and as a result, terminates transcription.[22]
dCas9 also proves to be efficient when it comes to altering certain proteins that can create diseases. When the dCas9 attaches to a form of RNA called guide-RNA, it prevents the proliferation of repeating codons and DNA sequences that might be harmful to an organism's genome. Essentially, when multiple repeat codons are produced, it elicits a response, or recruits an abundance of dCas9 to combat the overproduction of those codons and results in the shut-down of transcription. dCas9 works synergistically with gRNA and directly affects the DNA polymerase II from continuing transcription.
Further explanation of how the dCas9 protein works can be found in their utilization of plant genomes by the regulation of gene production in plants to either increase or decrease certain characteristics. The CRISPR-CAS9 system has the ability to either upregulate or downregulate genes. The dCas9 proteins are a component of the CRISPR-CAS9 system and these proteins can repress certain areas of a plant gene. This happens when dCAS9 binds to repressor domains, and in the case of the plants, deactivation of a regulatory gene such as AtCSTF64, does occur.[23]
Bacteria are another focus of the usage of dCas9 proteins as well. Since eukaryotes have a larger DNA makeup and genome; the much smaller bacteria are easy to manipulate. As a result, eukaryotes use dCas9 to inhibit RNA polymerase from continuing the process of transcription of genetic material.[24]
Structural and biochemical studies
Crystal structure
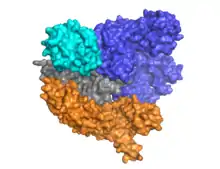
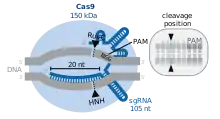
Cas9 features a bi-lobed architecture with the guide RNA nestled between the alpha-helical lobe (blue) and the nuclease lobe (cyan, orange, and gray). These two lobes are connected through a single bridge helix. There are two nuclease domains located in the multi-domain nuclease lobe, the RuvC (gray) which cleaves the non-target DNA strand, and the HNH nuclease domain (cyan) that cleaves the target strand of DNA. The RuvC domain is encoded by sequentially disparate sites that interact in the tertiary structure to form the RuvC cleavage domain (See right figure).
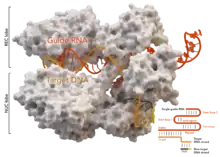
A key feature of the target DNA is that it must contain a protospacer adjacent motif (PAM) consisting of the three-nucleotide sequence- NGG. This PAM is recognized by the PAM-interacting domain (PI domain, orange) located near the C-terminal end of Cas9. Cas9 undergoes distinct conformational changes between the apo, guide RNA bound, and guide RNA:DNA bound states.
Cas9 recognizes the stem-loop architecture inherent in the CRISPR locus, which mediates the maturation of crRNA-tracrRNA ribonucleoprotein complex.[26] Cas9 in complex with CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) further recognizes and degrades the target dsDNA.[27] In the co-crystal structure shown here, the crRNA-tracrRNA complex is replaced by a chimeric single-guide RNA (sgRNA, in red) which has been proved to have the same function as the natural RNA complex.[4] The sgRNA base paired with target ssDNA is anchored by Cas9 as a T-shaped architecture. This crystal structure of the DNA-bound Cas9 enzyme reveals distinct conformational changes in the alpha-helical lobe with respect to the nuclease lobe, as well as the location of the HNH domain. The protein consists of a recognition lobe (REC) and a nuclease lobe (NUC). All regions except the HNH form tight interactions with each other and sgRNA-ssDNA complex, while the HNH domain forms few contacts with the rest of the protein. In another conformation of Cas9 complex observed in the crystal, the HNH domain is not visible. These structures suggest the conformational flexibility of HNH domain.
To date, at least three crystal structures have been studied and published. One representing a conformation of Cas9 in the apo state,[25] and two representing Cas9 in the DNA bound state.[28][1]
Interactions with sgRNA
In sgRNA-Cas9 complex, based on the crystal structure, REC1, BH and PI domains have important contacts with backbone or bases in both repeat and spacer region.[1][28] Several Cas9 mutants including REC1 or REC2 domains deletion and residues mutations in BH have been tested. REC1 and BH related mutants show lower or none activity compared with wild type, which indicate these two domains are crucial for the sgRNA recognition at repeat sequence and stabilization of the whole complex. Although the interactions between spacer sequence and Cas9 as well as PI domain and repeat region need further studies, the co-crystal demonstrates clear interface between Cas9 and sgRNA.
DNA cleavage
Previous sequence analysis and biochemical studies have posited that Cas9 contains two nuclease domains: an McrA-like HNH nuclease domain and a RuvC-like nuclease domain.[29] These HNH and RuvC-like nuclease domains are responsible for cleavage of the complementary/target and non-complementary/non-target DNA strands, respectively.[4] Despite low sequence similarity, the sequence similar to RNase H has a RuvC fold (one member of RNase H family) and the HNH region folds as T4 Endo VII (one member of HNH endonuclease family).
Wild-type S. pyogenes Cas9 requires magnesium (Mg2+) cofactors for the RNA-mediated DNA cleavage; however, Cas9 has been shown to exhibit varying levels of activity in the presence of other divalent metal ions.[4] For instance, Cas9 in the presence of manganese (Mn2+) has been shown to be capable of RNA-independent DNA cleavage.[30] The kinetics of DNA cleavage by Cas9 have been of great interest to the scientific community, as this data provides insight into the intricacies of the reaction. While the cleavage of DNA by RNA-bound Cas9 has been shown to be relatively rapid (k ≥ 700 s−1), the release of the cleavage products is very slow (t1/2 = ln(2)/k ≈ 43-91 h), essentially rendering Cas9 a single-turnover enzyme.[31] Additional studies regarding the kinetics of Cas9 have shown engineered Cas9 to be effective in reducing off-target effects by modifying the rate of the reaction.[32][33]
The cleavage efficiency of Cas9 depends on numerous factors. A key requirement is the presence of a valid PAM at the non-target strand 3 nucleotides downstream from the cleavage site.[34] The canonical PAM sequence for S. Pyogenes Cas9 is NGG, but alternative motifs are tolerated with lower cleavage activity. The most efficient alternative PAM motifs for the wild-type S. Pyogenes Cas9 are NAG and NGA.[35][36] The sequence composition at the target DNA site complementary to the 20 nucletode spacer region of the gRNA also affects cleavage efficiency. The most relevant nucleotide composition properties that impact efficiency are those in the PAM-proximal region.[37][38][36] Free energy changes of nucleic acids are also highly relevant in defining cleavage activity.[39] Guide RNAs that bind to the DNA forming a duplex that falls into a restricted range of binding free energy changes that excludes extremely weak or stable bindings generally perform efficiently.[36] Stable guide RNA folding conformations can also impair cleavage.[40]
Problems bacteria pose to Cas9 editing
Most archaea and bacteria stubbornly refuse to allow a Cas9 to edit their genome. This is because they can attach foreign DNA, that does not affect them, into their genome. Another way that these cells defy Cas9 is by process of restriction modification (RM) system. When a bacteriophage enters a bacteria or archaea cell it is targeted by the RM system. The RM system then cuts the bacteriophages DNA into separate pieces by restriction enzymes and uses endonucleases to further destroy the strands of DNA. This poses a problem to Cas9 editing because the RM system also targets the foreign genes added by the Cas9 process.[41]
Applications of Cas9 to transcription tuning
Interference of transcription by dCas9
Due to the unique ability of Cas9 to bind to essentially any complement sequence in any genome, researchers wanted to use this enzyme to repress transcription of various genomic loci. To accomplish this, the two crucial catalytic residues of the RuvC and HNH domain can be mutated to alanine abolishing all endonuclease activity of Cas9. The resulting protein coined 'dead' Cas9 or 'dCas9' for short, can still tightly bind to dsDNA. This catalytically inactive Cas9 variant has been used for both mechanistic studies into Cas9 DNA interrogative binding and as a general programmable DNA binding RNA-Protein complex.
The interaction of dCas9 with target dsDNA is so tight that high molarity urea protein denaturant can not fully dissociate the dCas9 RNA-protein complex from dsDNA target.[42] dCas9 has been targeted with engineered single guide RNAs to transcription initiation sites of any loci where dCas9 can compete with RNA polymerase at promoters to halt transcription.[43] Also, dCas9 can be targeted to the coding region of loci such that inhibition of RNA Polymerase occurs during the elongation phase of transcription.[43] In Eukaryotes, silencing of gene expression can be extended by targeting dCas9 to enhancer sequences, where dCas9 can block assembly of transcription factors leading to silencing of specific gene expression.[10] Moreover, the guide RNAs provided to dCas9 can be designed to include specific mismatches to its complementary cognate sequence that will quantitatively weaken the interaction of dCas9 for its programmed cognate sequence allowing a researcher to tune the extent of gene silencing applied to a gene of interest.[43] This technology is similar in principle to RNAi such that gene expression is being modulated at the RNA level. However, the dCas9 approach has gained much traction as there exist less off-target effects and in general larger and more reproducible silencing effects through the use of dCas9 compared to RNAi screens.[44] Furthermore, because the dCas9 approach to gene silencing can be quantitatively controlled, a researcher can now precisely control the extent to which a gene of interest is repressed allowing more questions about gene regulation and gene stoichiometry to be answered.
Beyond direct binding of dCas9 to transcriptionally sensitive positions of loci, dCas9 can be fused to a variety of modulatory protein domains to carry out a myriad of functions. Recently, dCas9 has been fused to chromatin remodeling proteins (HDACs/HATs) to reorganize the chromatin structure around various loci.[43] This is important in targeting various eukaryotic genes of interest as heterochromatin structures hinder Cas9 binding. Furthermore, because Cas9 can react to heterochromatin, it is theorized that this enzyme can be further applied to studying the chromatin structure of various loci.[43] Additionally, dCas9 has been employed in genome wide screens of gene repression. By employing large libraries of guide RNAs capable of targeting thousands of genes, genome wide genetic screens using dCas9 have been conducted.[45]
Another method for silencing transcription with Cas9 is to directly cleave mRNA products with the catalytically active Cas9 enzyme.[46] This approach is made possible by hybridizing ssDNA with a PAM complement sequence to ssRNA allowing for a dsDNA-RNA PAM site for Cas9 binding. This technology makes available the ability to isolate endogenous RNA transcripts in cells without the need to induce chemical modifications to RNA or RNA tagging methods.
Transcription activation by dCas9 fusion proteins
In contrast to silencing genes, dCas9 can also be used to activate genes when fused to transcription activating factors.[43] These factors include subunits of bacterial RNA Polymerase II and traditional transcription factors in eukaryotes. Recently, genome-wide screens of transcription activation have also been accomplished using dCas9 fusions named 'CRISPRa' for activation.[45]
See also
- Cas9.com
- CRISPR
- CRISPR gene editing
- Genome editing
- Zinc finger nuclease
- Transcription activator-like effector nuclease
References
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O (February 2014). "Crystal structure of Cas9 in complex with guide RNA and target DNA". Cell. 156 (5): 935–49. doi:10.1016/j.cell.2014.02.001. PMC 4139937. PMID 24529477.
- "The Nobel Prize in Chemistry 2020". NobelPrize.org. Retrieved 2020-10-07.
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (March 2011). "CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III". Nature. 471 (7340): 602–607. Bibcode:2011Natur.471..602D. doi:10.1038/nature09886. PMC 3070239. PMID 21455174.
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (August 2012). "A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity". Science. 337 (6096): 816–21. Bibcode:2012Sci...337..816J. doi:10.1126/science.1225829. PMC 6286148. PMID 22745249.
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA (March 2015). "Cas9 specifies functional viral targets during CRISPR-Cas adaptation". Nature. 519 (7542): 199–202. Bibcode:2015Natur.519..199H. doi:10.1038/nature14245. PMC 4385744. PMID 25707807.
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. (March 2007). "CRISPR provides acquired resistance against viruses in prokaryotes". Science. 315 (5819): 1709–12. Bibcode:2007Sci...315.1709B. doi:10.1126/science.1138140. hdl:20.500.11794/38902. PMID 17379808. S2CID 3888761.
- Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S (November 2010). "The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA". Nature. 468 (7320): 67–71. Bibcode:2010Natur.468...67G. CiteSeerX 10.1.1.451.9645. doi:10.1038/nature09523. PMID 21048762. S2CID 205222849.
- Mali P, Esvelt KM, Church GM (October 2013). "Cas9 as a versatile tool for engineering biology". Nature Methods. 10 (10): 957–63. doi:10.1038/nmeth.2649. PMC 4051438. PMID 24076990.
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM (September 2013). "CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering". Nature Biotechnology. 31 (9): 833–8. doi:10.1038/nbt.2675. PMC 3818127. PMID 23907171.
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS (July 2013). "CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes". Cell. 154 (2): 442–51. doi:10.1016/j.cell.2013.06.044. PMC 3770145. PMID 23849981.
- Esvelt KM, Smidler AL, Catteruccia F, Church GM (July 2014). "Concerning RNA-guided gene drives for the alteration of wild populations". eLife. 3. doi:10.7554/eLife.03401. PMC 4117217. PMID 25035423.
- Cyranoski D, Reardon S (22 April 2015). "Chinese scientists genetically modify human embryos". Nature. doi:10.1038/nature.2015.17378. S2CID 87604469.
- Doudna JA, Mali P (2016). CRISPR-Cas: a laboratory manual. Cold Spring Harbor, New York. ISBN 9781621821304. OCLC 922914104.
- Chen W, Page-McCaw PS (March 2019). "CRISPR/Cas9 gene editing". AccessScience. McGraw-Hill Education. doi:10.1036/1097-8542.168060.
- Ebina H, Misawa N, Kanemura Y, Koyanagi Y (2013-08-26). "Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus". Scientific Reports. 3 (1): 2510. Bibcode:2013NatSR...3E2510E. doi:10.1038/srep02510. PMC 3752613. PMID 23974631.
- Li H, Sheng C, Wang S, Yang L, Liang Y, Huang Y, Liu H, Li P, Yang C, Yang X, Jia L, Xie J, Wang L, Hao R, Du X, Xu D, Zhou J, Li M, Sun Y, Tong Y, Li Q, Qiu S, Song H (2017-03-22). "Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9". Frontiers in Cellular and Infection Microbiology. 7: 91. doi:10.3389/fcimb.2017.00091. PMC 5360708. PMID 28382278.
- Deveau H, Garneau JE, Moineau S (2010-10-13). "CRISPR/Cas system and its role in phage-bacteria interactions". Annual Review of Microbiology. Annual Reviews. 64 (1): 475–493. doi:10.1146/annurev.micro.112408.134123. PMID 20528693.
- Horvath P, Barrangou R (January 2010). "CRISPR/Cas, the immune system of bacteria and archaea". Science. 327 (5962): 167–70. Bibcode:2010Sci...327..167H. doi:10.1126/science.1179555. PMID 20056882. S2CID 17960960.
- Karginov FV, Hannon GJ (January 2010). "The CRISPR system: small RNA-guided defense in bacteria and archaea". Molecular Cell. 37 (1): 7–19. doi:10.1016/j.molcel.2009.12.033. PMC 2819186. PMID 20129051.
- Jensen ED, Ferreira R, Jakočiūnas T, Arsovska D, Zhang J, Ding L, et al. (March 2017). "Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies". Microbial Cell Factories. 16 (1): 46. doi:10.1186/s12934-017-0664-2. PMC 5353793. PMID 28298224.
- Pinto BS, Saxena T, Oliveira R, Méndez-Gómez HR, Cleary JD, Denes LT, McConnell O, Arboleda J, Xia G, Swanson MS, Wang ET (November 2017). "Impeding Transcription of Expanded Microsatellite Repeats by Deactivated Cas9". Molecular Cell. 68 (3): 479–490.e5. doi:10.1016/j.molcel.2017.09.033. PMC 6013302. PMID 29056323.
- O'Geen H, Ren C, Nicolet CM, Perez AA, Halmai J, Le VM, Mackay JP, Farnham PJ, Segal DJ (September 2017). "dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression". Nucleic Acids Research. 45 (17): 9901–9916. doi:10.1093/nar/gkx578. PMC 5622328. PMID 28973434.
- Lowder LG, Paul JW, Qi Y (2017). "Multiplexed Transcriptional Activation or Repression in Plants Using CRISPR-dCas9-Based Systems". Plant Gene Regulatory Networks. Methods in Molecular Biology. Vol. 1629. pp. 167–184. doi:10.1007/978-1-4939-7125-1_12. ISBN 978-1-4939-7124-4. PMID 28623586.
- Barrangou R, Horvath P (June 2017). "A decade of discovery: CRISPR functions and applications". Nature Microbiology. 2 (7): 17092. doi:10.1038/nmicrobiol.2017.92. PMID 28581505. S2CID 19072900.
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA (March 2014). "Structures of Cas9 endonucleases reveal RNA-mediated conformational activation". Science. 343 (6176): 1247997. doi:10.1126/science.1247997. PMC 4184034. PMID 24505130.
- Wiedenheft B, Sternberg SH, Doudna JA (February 2012). "RNA-guided genetic silencing systems in bacteria and archaea". Nature. 482 (7385): 331–8. Bibcode:2012Natur.482..331W. doi:10.1038/nature10886. PMID 22337052. S2CID 205227944.
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (November 2013). "Genome engineering using the CRISPR-Cas9 system". Nature Protocols. 8 (11): 2281–2308. doi:10.1038/nprot.2013.143. PMC 3969860. PMID 24157548.
- Anders C, Niewoehner O, Duerst A, Jinek M (September 2014). "Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease". Nature. 513 (7519): 569–73. Bibcode:2014Natur.513..569A. doi:10.1038/nature13579. PMC 4176945. PMID 25079318.
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (March 2006). "A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action". Biology Direct. 1 (1): 7. doi:10.1186/1745-6150-1-7. PMC 1462988. PMID 16545108.
- Sundaresan R, Parameshwaran HP, Yogesha SD, Keilbarth MW, Rajan R (December 2017). "RNA-Independent DNA Cleavage Activities of Cas9 and Cas12a". Cell Reports. 21 (13): 3728–3739. doi:10.1016/j.celrep.2017.11.100. PMC 5760271. PMID 29281823.
- Raper AT, Stephenson AA, Suo Z (February 2018). "Functional Insights Revealed by the Kinetic Mechanism of CRISPR/Cas9". Journal of the American Chemical Society. 140 (8): 2971–2984. doi:10.1021/jacs.7b13047. PMID 29442507.
- Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, et al. (August 2018). "Directed evolution of CRISPR-Cas9 to increase its specificity". Nature Communications. 9 (1): 3048. Bibcode:2018NatCo...9.3048L. doi:10.1038/s41467-018-05477-x. PMC 6078992. PMID 30082838.
- Singh D, Wang Y, Mallon J, Yang O, Fei J, Poddar A, et al. (April 2018). "Mechanisms of improved specificity of engineered Cas9s revealed by single-molecule FRET analysis". Nature Structural & Molecular Biology. 25 (4): 347–354. doi:10.1038/s41594-018-0051-7. PMC 6195204. PMID 29622787.
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (August 2012). "A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity". Science. 337 (6096): 816–21. doi:10.1126/science.1225829. PMC 6286148. PMID 22745249.
- Kim N, Kim HK, Lee S, et al. (2020). "Prediction of the sequence-specific cleavage activity of Cas9 variants". Nat Biotechnol. 38: 1328–1336. doi:10.1038/s41587-020-0537-9. PMID 32514125.
- Corsi GI, Qu K, Alkan F, Pan X, Luo Y, Gorodkin J (May 2022). "CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context". Nature Communications. 13 (3006). doi:10.1038/s41467-022-30515-0. PMC 9151727. PMID 35637227.
- Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, Wu D, Cong L, Zhang F, Liu JS, Brown M, Liu XS (Aug 2015). "Sequence determinants of improved CRISPR sgRNA design". Genome Res. 25 (8). doi:10.1101/gr.191452.115. PMC 4509999. PMID 26063738.
- Xiang X, Corsi, GI, Anthon C, Qu K, Pan X, Liang X, Han P, Dong Z, Liu L, Zhong J, Ma T, Wang J, Zhang X, Jiang H, Xu F, Liu X, Xu X, Wang J, Yang H, Bolund L, Church GM, Lin L, Gorodkin J, Luo Y (May 2021). "Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning". Nature Communications. 12 (1): 3238. doi:10.1038/s41467-021-23576-0. PMC 8163799. PMID 34050182.
- Alkan F, Wenzel A, Anthon C, Havgaard JH, Gorodkin J (October 2018). "CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters". Genome Biology. 19 (177): 177. doi:10.1186/s13059-018-1534-x. PMC 6203265. PMID 30367669.
- Thyme SB, Akhmetova L, Montague TG, Valen E, Schier AF (Jun 2016). "Internal guide RNA interactions interfere with Cas9-mediated cleavage". Nature Communications. 7 (11750). doi:10.1038/ncomms11750. PMC 4906408. PMID 27282953.
- Kusano K, Naito T, Handa N, Kobayashi I (November 1995). "Restriction-modification systems as genomic parasites in competition for specific sequences". Proceedings of the National Academy of Sciences of the United States of America. 92 (24): 11095–9. Bibcode:1995PNAS...9211095K. doi:10.1073/pnas.92.24.11095. PMC 40578. PMID 7479944.
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (March 2014). "DNA interrogation by the CRISPR RNA-guided endonuclease Cas9". Nature. 507 (7490): 62–7. Bibcode:2014Natur.507...62S. doi:10.1038/nature13011. PMC 4106473. PMID 24476820.
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA (August 2013). "Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system". Nucleic Acids Research. 41 (15): 7429–37. doi:10.1093/nar/gkt520. PMC 3753641. PMID 23761437.
- Heintze J, Luft C, Ketteler R (2013). "A CRISPR CASe for high-throughput silencing". Frontiers in Genetics. 4: 193. doi:10.3389/fgene.2013.00193. PMC 3791873. PMID 24109485.
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS (October 2014). "Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation". Cell. 159 (3): 647–61. doi:10.1016/j.cell.2014.09.029. PMC 4253859. PMID 25307932.
- O'Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA (December 2014). "Programmable RNA recognition and cleavage by CRISPR/Cas9". Nature. 516 (7530): 263–6. Bibcode:2014Natur.516..263O. doi:10.1038/nature13769. PMC 4268322. PMID 25274302.
Further reading
- Kennedy EM, Cullen BR (May 2015). "Bacterial CRISPR/Cas DNA endonucleases: A revolutionary technology that could dramatically impact viral research and treatment". Virology. 479–480: 213–20. doi:10.1016/j.virol.2015.02.024. PMC 4424069. PMID 25759096.
- Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, et al. (May 2020). "Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza". Cell. 181 (4): 865–876.e12. doi:10.1016/j.cell.2020.04.020. PMC 7189862. PMID 32353252.
- Steven Levy (March 18, 2020). "Could Crispr Be Humanity's Next Virus Killer?". Wired.
External links
- Overview of all the structural information available in the PDB for UniProt: Q99ZW2 (Streptococcus pyogenes CRISPR-associated endonuclease Cas9/Csn1) at the PDBe-KB.