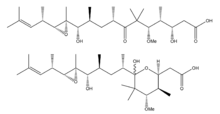
Gephyronic acid
 | |
| Names | |
|---|---|
| IUPAC name
3,11,12,13-tetrahydroxy-5-methoxy-4,6,6,8,10,12,14,16-octamethyl-7-oxoheptadec-15-enoic acid | |
| Identifiers | |
3D model (JSmol) |
|
| ChemSpider | |
PubChem CID |
|
InChI
| |
SMILES
| |
| Properties | |
Chemical formula |
C26H48O8 |
| Molar mass | 488.660 g/mol |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
Gephyronic acid is a polyketide that exists as an equilibrating mixture of structural isomers. In nature, gephyronic acid is produced by slow growing myxobacterium: Archangium gephyra strain Ar3895 and Cystobacter violaceus strain Cb vi76.[1] It is the first antibiotic in myxobacteria that was reported to specifically inhibit eukaryotic protein synthesis.
Biological properties
Preliminary studies demonstrated that gephyronic acid inhibited growth of yeast and molds as well as elicited a cytostatic effect through the inhibition of eukaryotic protein synthesis in mammalian cell cultures. Feeding experiments done with radioactive precursors showed a drastic difference in incorporation of leucine by a human leukemic cell like K-562, but little difference in the incorporation of uridine and thymidine.[2] This suggested that the primary target of gephyronic acid is protein synthesis. As such, it is a potential target for cancer chemotherapy. Gene expression profiling of human breast cancer cell lines is underway in an effort to further define the potential of gephyronic acid as a chemotherapeutic lead.[3]
Screening of a library of compounds derived from myxobacteria found that gephyronic acid was the strongest inhibitor of processing bodies (P-bodies) assembly.[4] P-bodies are discrete cytoplasmic mRNP granules that contain non-translating mRNA and protein from the mRNA decay pathway and from miRNA silencing machinery. Within P-bodies, mRNAs can be degraded, but components of P-bodies can rapidly cycle in and out to return to translation.[4] The mechanism of P-body assembly inhibition by gephyronic acid has not been characterized, but initial studies suggest that the mode of action could be through stalling ribosomes on mRNA or by reflecting early steps of translation initiation, such as binding of ribosomal subunits or initiation factors.
The same study also found that gephyronic acid inhibits eIF2α-phosphorylation and formation of stress granules under stress conditions. Stress granules contain non-translating mRNAs and translation initiation factors, suggesting that they may form as a result of aggregation of mRNPs stalled during translation initiation.[4] By monitoring immunofluresence of an established stress granules marker, it was found that stress granule formation was inhibited in the presence of gephyronic acid. Gephyronic acid may have a direct or indirect effect on translation initiation factor eIF2α, which would trap mRNA into nonfunctional initiation complexes, inhibiting both P-body and stress granule formation.[4]
Biosynthesis
Sequencing of the PKS gene cluster in C. violaceus was conducted and confirmed to reveal five type I polyketide synthases and post-PKS tailoring enzymes with O-methyltransferase and cytochrome P450 monooxygenase.[5] The overall structure correlates well with modular arrangement of PKS-encoded proteins, aside from some unexpected elements that are likely caused by inactive domains. Initial loading uses a GCN5-related N-acetyltransferase (GNAT) domain instead of the typical AT domain.
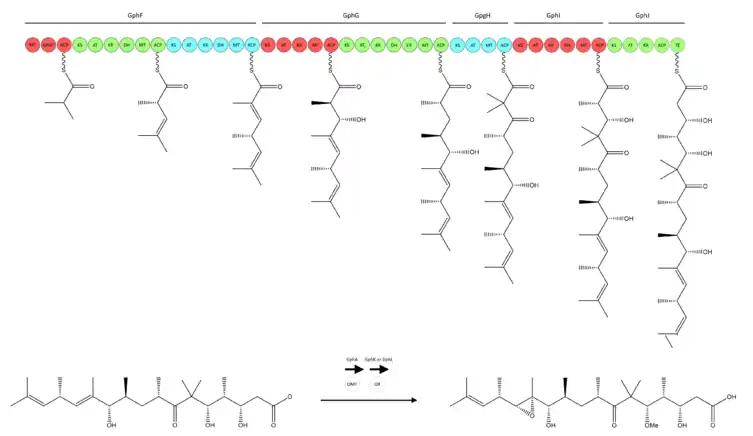

Gephyronic acid contains a methyl ether at C-5 and a C-12/C-13 epoxide. These functional groups are incorporated by post-PKS tailoring enzymes. GphA is likely responsible for installation of the C-5 methyl ether. O-methyltransferases SpiB and SpiK used in spirangien biosynthesis exhibit the same SAM-binding motif as GphA.[6]

GphK is a member of the cytochrome p450 superfamily and is suspected to carry out the epoxidation of the C12-C13 olefin. Such epoxidation in post-PKS modifications has been seen in epothilone biosynthesis by EpoK.[2] In EpoK, the consensus mechanism of epoxidation by P450 involves the formation of a pi-complex between an oxoferryl pi-cation radical species (FeIV) and the olefin pi bond, followed by electron transfer, formation of the olefin pi-cation radical and finally epoxidation.[7]
However, it is also possible that in addition to cytochrome p450, a FAD-dependent monoxygenase is also required to install the epoxide. This codependent process is seen in tirandamycin biosynthesis by TamL.[2] Experiments to clarify the function of these enzymes in gephyronic acid biosynthesis are underway.[5]
References
- Wenzel, Silke C.; Müller, Rolf (21 May 2009). "Myxobacteria—'microbial factories' for the production of bioactive secondary metabolites". Molecular BioSystems. 5 (6): 567–74. doi:10.1039/b901287g. ISSN 1742-2051. PMID 19462013.
- Sasse, F.; Steinmetz, H.; Höfle, G.; Reichenbach, H. (1 January 1995). "Gephyronic acid, a novel inhibitor of eukaryotic protein synthesis from Archangium gephyra (myxobacteria). Production, isolation, physico-chemical and biological properties, and mechanism of action". The Journal of Antibiotics. 48 (1): 21–25. doi:10.7164/antibiotics.48.21. ISSN 0021-8820. PMID 7868385.
- Young, Jeanette; Leliaert, Amy; Schafer, Zachary T.; Taylor, Richard E. (28 October 2014). "Abstract C67: Gene expression profiling with human breast cancer cell lines exposed to gephyronic acid". Cancer Research. 71 (18 Supplement): C67. doi:10.1158/1538-7445.fbcr11-c67.
- Martínez, Javier P.; Pérez-Vilaró, Gemma; Muthukumar, Yazh; Scheller, Nicoletta; Hirsch, Tatjana; Diestel, Randi; Steinmetz, Heinrich; Jansen, Rolf; Frank, Ronald (1 November 2013). "Screening of small molecules affecting mammalian P-body assembly uncovers links with diverse intracellular processes and organelle physiology". RNA Biology. 10 (11): 1661–1669. doi:10.4161/rna.26851. ISSN 1547-6286. PMC 3907476. PMID 24418890.
- Young, Jeanette; Stevens, D. Cole; Carmichael, Rory; Tan, John; Rachid, Shwan; Boddy, Christopher N.; Müller, Rolf; Taylor, Richard E. (27 December 2013). "Elucidation of Gephyronic Acid Biosynthetic Pathway Revealed Unexpected SAM-Dependent Methylations". Journal of Natural Products. 76 (12): 2269–2276. doi:10.1021/np400629v. ISSN 0163-3864. PMID 24298873.
- Clarke, Steven (5 February 2002). "The methylator meets the terminator". Proceedings of the National Academy of Sciences. 99 (3): 1104–1106. Bibcode:2002PNAS...99.1104C. doi:10.1073/pnas.042004099. ISSN 0027-8424. PMC 122150. PMID 11830650.
- Kells, Petrea M.; Ouellet, Hugues; Santos-Aberturas, Javier; Aparicio, Jesus F.; Podust, Larissa M. (27 August 2010). "Structure of Cytochrome P450 PimD Suggests Epoxidation of the Polyene Macrolide Pimaricin Occurs via a Hydroperoxoferric Intermediate". Chemistry & Biology. 17 (8): 841–851. doi:10.1016/j.chembiol.2010.05.026. PMC 2932657. PMID 20797613.