Killer activation receptor
Killer Activation Receptors (KARs) are receptors expressed on the plasmatic membrane of Natural Killer cells (NK cells). KARs work together with inhibitory receptors (abbreviated as KIRs in the text), which inactivate them in order to regulate the NK cells functions on hosted or transformed cells. These two kinds of specific receptors have some morphological features in common, such as being transmembrane proteins. The similarities are specially found in the extracellular domains and, the differences tend to be in the intracellular domains. KARs and KIRs can have tyrosine containing activatory or inhibitory motifs in the intracellular part of the receptor molecule (they are called ITAMs and ITIMs).
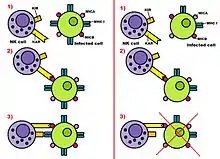
At first, it was thought that there were only one KAR and one KIR (two-receptor model). In the last decade, many different KARs and KIRs, such as NKp46 or NKG2D, have been discovered (opposing-signals model). NKG2D is activated by the cell-surface ligands MICA and ULBP2.[1]
There is an unfortunate confusion about the KIR acronym. The KIR term has been started to be being used parallelly both for the Killer-cell immunoglobulin-like receptors (KIRs) and for the Killer Inhibitory Receptors. The Killer-cell immunoglobulin-like receptors involve both activatory and inhibitory receptors.[2] Killer-cell inhibitory receptors involve both immunoglobulin-like receptors and C-type lectin-like receptors.[3]
Morphology
There are two different kinds of surface receptors which are responsible for triggering NK-mediated natural cytotoxicity: the NK KARs (meaning: Killer Activation Receptors) and the NK KIRs (meaning: Killer Inhibitory Receptors). Such receptors have a broad binding specificity and, therefore, are able to broadcast opposite signals. It is the balance between these competing signals that determines whether or not the cytotoxic activity of the NK cell should get started.
As KARs and KIRs are receptors with antagonic effects on NK cells, they have some structural characteristics in common. Firstly, both of them are usually transmembrane proteins. Apart from that, the extracellular domains of these proteins tend to have similar molecular features and are responsible for ligand recognition.
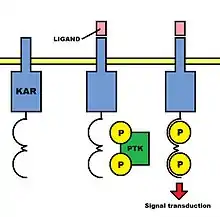
Therefore, the opposing functions these receptors have must be attributed to differences in their intracellular domains. KARs proteins possess positively charged transmembrane residues and short cytoplasmic tails that contain few intracellular signaling domains. In contrast, KIRs proteins usually have long cytoplasmic tails.
As the chains which form KARs are not able to mediate any signal transduction in isolation, a common feature of such receptors is the presence of noncovalently linked subunits that contain immunoreceptor tyrosine-based activation motifs (ITAMs) in their cytoplasmic tails. ITAMs are composed of a conserved sequence of amino acids, including two Tyr-x-x-Leu/Ile elements (where x is any amino acid) separated by six to eight amino acid residues. When the binding of an activation ligand to an activation receptor complex occurs, the tyrosine residues in the ITAMs in the associated chain are phosphorylated by kinases, and a signal that promotes natural cytotoxicity is conveyed to the interior of the NK cell. Therefore, ITAMs are involved in the facilitation of signal transduction. These subunits are moreover composed of an accessory signaling molecule such as CD3ζ, the γc chain, or one of two adaptor proteins called DAP10 and DAP12. All of these molecules possess negatively charged transmembrane domains.
A common feature of members of all KIR is the presence of immunoreceptor tyrosine-based inhibition motifs (ITIMs) in their cytoplasmic tails. ITIMs are composed of the sequence Ile/Val/Leu/Ser-x-Tyr-x-x-Leu/Val, where x denotes any amino acid. The latter are essential to the signaling functions of these molecules. When an inhibitory receptor is stimulated by the binding of MHC class I, kinases and phosphatases are recruited to the receptor complex. This is how ITIMs counteract the effect of kinases initiated by activating receptors and manage to inhibit the signal transduction within the NK cell.
Types
Following the criterion of the structure, KARs are found in three different groups. The first group of receptors where KARs are included is called Natural Cytotoxicity Receptors (NCR), and it only includes activation receptors. The two other classes are: Natural Killer Group 2 (NKG2), which includes activation and inhibition receptors (in this article, we will only describe NKG2 with an activator role), and some KIRs which, exceptionally, don’t have an inhibitor role.
The three receptors which are included in the NCR class are NKp46, NKp44 and NKp30. The crystal structure of NKp46, which is representative for all three NCR, has been determined. It has two C2-set immunoglobulin domains, and it’s probable that the binding site for its ligand is near the interdomain hinge.
Two NKG2-class receptors are NKG2D, considered the most important and the better studied NKG2 receptor, and CD94/NKG2C. NKG2D, which doesn’t bind to CD94, is a homodimeric lectin-like receptor. On the other hand, CD94/NKG2C consists in a complex formed by the CD94 protein, which is a C-type lectin molecule bonded to the NKG2C protein. This molecule can bind to five classes of NKG2 (A, B, C, E and H), but the union can trigger an activation or an inhibition response, depending on the NKG2 molecule (CD94/NKG2A, for example, is an inhibitor complex).
It’s important to mention that most KIRs have an inhibitor function, which has been generalized in this article, but a few KIRs that have an activator role also exist. One of these activatory KIRs is KIR2DS1, which has an Ig-like structure, like KIRs in general.
Finally, there is CD16, a low affinity Fc receptor (FcγRIII) which contains N-glycosylation sites; therefore, it is a glycoprotein.
Killer Activation Receptors are associated with signaling intracellular chains. In fact, these intracellular domains determine the opposite functions of activation and inhibitory receptors. Activation receptors are associated with an accessory signaling molecule (for instance, CD3ζ) or with an adaptor protein, which can be either DAP10 or DAP12. All of these signaling molecules contain immunoreceptor tyrosine-based activated motifs (ITAMs), which are phosphorylated and consequently facilitate signal transduction.
Each of these receptors has a specific ligand, although some receptors that belong to the same class, such as NCR, recognize similar molecules.
Physiology
As we mentioned previously, natural killer cells can discharge their function properly through two types of receptors: Killer Activation Receptor (KAR) and Killer Inhibition Receptors (KIRs). Both type of receptors act together to activate or not activate the Natural Killer cell following the opposing-signals model.
KARs can detect a specific type of molecules: MICA and MICB. These molecules are in MHC class I of human cells and they are related to cellular stress: this is why MICA and MICB appear in infected or transformed cells but they aren't very common in healthy cells. KARs recognise MICA and MICB when they are in a huge proportion and get engaged. This engagement activates the natural killer cell to attack the transformed or infected cells. This action can be done in different ways. NK can kill directly the hosted cell, it can do it by segregating cytokines, IFN-β and IFN-α, or by doing both things.
Furthermore, there are other less common ligands, like carbohydrate domains, which are recognised by a group of receptors: C-type lectins (so named because they have calcium-dependent carbohydrates recognition domains).
In addition to lectins, there are other molecules implicated in the activation of NK. These additional proteins are: CD2 and CD16. The last one works in antibody-mediated recognition.
Finally, there is a group of proteins which are related to the activation in an unknown way. These are NKp30, Nkp44 and Nkp46.
To recapitulate, these ligands activate the NK, as we explained. However, before the activation, Killer Inhibition Receptors (KIRs) recognize certain molecules in the MHC class I of the hosted cell and get engaged with them. These molecules are typical of healthy cells but some of these molecules are repressed in infected or transformed cells. For this reason when the hosted cell is really infected the proportion of KARs engaged with ligands is bigger than the proportion of KIRs engaged with MHC I molecules. When this happens the NK is activated and the hosted cell is destroyed. On the other hand, if there are more KIRs engaged with MHC class I molecules than KARs engaged with ligands, the NK isn't activated and the suspicious hosted cell remains alive. To conclude, we should add that each KAR has its specific KIR and they always work together. For instance, the C-type lectins receptors are inhibited by some CD94/NKG2 complex.
KARs and KIRs: their role in cancer
One way by which NK cells are able to distinguish between normal and infected or transformed cells is by monitoring the amount of MHC class I molecules cells have on their surface provided both in an infected and in a tumor cell the expression of MHC class I decreases.
What happens in malignant transformations, that is to say cancers, is that a Killer Activation Receptor (KAR), located on the surface of the NK cell, binds to certain molecules which only appear on cells that are undergoing stress situations. In humans, this KAR is called NKG2D and the molecules it recognizes MICA and MICB. This binding provides a signal which induces the NK cell to kill the target cell.
Then, Killer Inhibitory Receptors (KIRs) examine the surface of the tumor cell in order to determine the levels of MHC class I molecules it has. If KIRs bind sufficiently to MHC class I molecules, the “killing signal” is overridden to prevent the killing of the cell. In contrast to this, if KIRs are not sufficiently engaged to MHC class I molecules, killing of the target cell proceeds.
References
- Song P, Zhao Q, Zou M (2020). "Targeting senescent cells to attenuate cardiovascular disease progression". Ageing Research Reviews. 60: 101072. doi:10.1016/j.arr.2020.101072. PMC 7263313. PMID 32298812.
- Parham, Peter (March 2004). "Killer cell immunoglobulin-like receptor diversity: balancing signals in the natural killer cell response". Immunology Letters. 92 (1–2): 11–13. doi:10.1016/j.imlet.2003.11.016. PMID 15081521.
- Radaev, Sergei; Sun, Peter D. (June 2003). "Structure and Function of Natural Killer Cell Surface Receptors". Annual Review of Biophysics and Biomolecular Structure. 32 (1): 93–114. doi:10.1146/annurev.biophys.32.110601.142347. PMID 12471063.
Further reading
- Richard, A.GOLDSBY; KINDT, Thomas J.; et al. (2003). Inmunology (5th ed.). New york: freeman. pp. 331–3. ISBN 0-7167-4947-5.
- ROITT. S (2008). Inmunologia Fundamentos (11th ed.). Buenos Aires: ; Editorial Médica panamericana. pp. 94–95. ISBN 978-950-06-0899-2.
- Murphy, Kenneth P.; Murphy, Kenneth M.; Travers, Paul; Walport, Mark; Janeway, Charles; Ehrenstein, Michael (2008). Janeway's Immunobiology. Garland Science. ISBN 978-0-8153-4123-9.
- Yokoyama, Wayne M. (2008). "Natural Killer Cells". In Paul, William E. (ed.). Fundamental Immunology. Lippincott Williams & Wilkins. pp. 483–517. ISBN 978-0-7817-6519-0.
- Doan, Thao; Celada Cotarelo, Antonio; Ovid Technologies (2013). Inmunología (in Spanish). Wolters Kluwer Health/Lippincott Williams & Wilkins. pp. 38–42. OCLC 932805424.
- Abbas, Abul K; Lichtman, Andrew H; Baker, David L; Baker, Alexandra (2005). Cellular and molecular immunology. Elsevier Saunders. ISBN 978-1-4160-2389-0. OCLC 981398164.
- Natural killer and leucocyte receptor complexes. Munksgaard. 2001. pp. 53, 115 and 123. OCLC 248460612.
- TAK W., Mak; SAUNDERS, Mary (2010). Primer to the Immune Response: Academic Cell Update Edition. Academic Press. pp. 174, 176, 379, 415, 431 and 437. ISBN 978-0-12-384743-0.
- DOAN, Thao; MELVOLD, Roger (2005). Concise medical immunology. Lippincott Williams & Wilkins. pp. 32–34. ISBN 978-0-7817-5741-6.
- Sentman, Charles L.; Barber, Melissa A.; Barber, Amorette; Zhang, Tong (2006). "NK Cell Receptors as Tools in Cancer Immunotherapy". Advances in Cancer Research. Vol. 95. pp. 249–292. doi:10.1016/S0065-230X(06)95007-6. ISBN 9780120066957. PMID 16860660.
- Sullivan, Lucy C.; Clements, Craig S.; Beddoe, Travis; Johnson, Darryl; Hoare, Hilary L.; Lin, Jie; Huyton, Trevor; Hopkins, Emma J.; Reid, Hugh H.; Wilce, Matthew C.J.; Kabat, Juraj; Borrego, Francisco; Coligan, John E.; Rossjohn, Jamie; Brooks, Andrew G. (December 2007). "The Heterodimeric Assembly of the CD94-NKG2 Receptor Family and Implications for Human Leukocyte Antigen-E Recognition". Immunity. 27 (6): 900–911. doi:10.1016/j.immuni.2007.10.013. PMID 18083576.
- Foster, Christine E.; Colonna, Marco; Sun, Peter D. (14 November 2003). "Crystal Structure of the Human Natural Killer (NK) Cell Activating Receptor NKp46 Reveals Structural Relationship to Other Leukocyte Receptor Complex Immunoreceptors". Journal of Biological Chemistry. 278 (46): 46081–46086. doi:10.1074/jbc.M308491200. PMID 12960161.
- Hibbs, ML; Classon, BJ; Walker, ID; McKenzie, IF; Hogarth, PM (1988). "The structure of the murine Fc receptor for IgG. Assignment of intrachain disulfide bonds, identification of N-linked glycosylation sites, and evidence for a fourth form of Fc receptor". Journal of Immunology. 140 (2): 544–50. PMID 2961814.
- Agüera-González, Sonia; Gross, Catharina C.; Fernández-Messina, Lola; Ashiru, Omodele; Esteso, Gloria; Hang, Howard C.; Reyburn, Hugh T.; Long, Eric O.; Valés-Gómez, Mar (December 2011). "Palmitoylation of MICA, a ligand for NKG2D, mediates its recruitment to membrane microdomains and promotes its shedding". European Journal of Immunology. 41 (12): 3667–3676. doi:10.1002/eji.201141645. PMC 3709245. PMID 21928280.
- Bolanos, Fred D.; Tripathy, Sandeep K. (1 March 2011). "Activation Receptor-Induced Tolerance of Mature NK Cells In Vivo Requires Signaling through the Receptor and Is Reversible". The Journal of Immunology. 186 (5): 2765–2771. doi:10.4049/jimmunol.1003046. PMC 3256587. PMID 21263069.
- López-Botet, M. "Activacion Celulas NK". Archived from the original on 8 August 2011. Retrieved 8 November 2011.