RNA therapeutics
RNA therapeutics are a new class of medications based on ribonucleic acid (RNA). Research has been working on clinical use since the 1990s, with significant success in cancer therapy in the early 2010s.[1] In 2020 and 2021, mRNA vaccines have been developed globally for use in combating the coronavirus disease (COVID-19 pandemic).[2] The Pfizer–BioNTech COVID-19 vaccine was the first mRNA vaccine approved by a medicines regulator, followed by the Moderna COVID-19 vaccine, and others.
The main types of RNA therapeutics are those based on messenger RNA (mRNA), antisense RNA (asRNA), RNA interference (RNAi), and RNA aptamers. Of the four types, mRNA-based therapy is the only type which is based on triggering synthesis of proteins within cells, making it particularly useful in vaccine development.[3] Antisense RNA is complementary to coding mRNA and is used to trigger mRNA inactivation to prevent the mRNA from being used in protein translation.[4] RNAi-based systems use a similar mechanism, and involve the use of both small interfering RNA (siRNA) and micro RNA (miRNA) to prevent mRNA translation.[5][6] However, RNA aptamers are short, single stranded RNA molecules produced by directed evolution to bind to a variety of biomolecular targets with high affinity thereby affecting their normal in vivo activity.[7][8][9]
RNA is synthesized from template DNA by RNA polymerase with messenger RNA (mRNA) serving as the intermediary biomolecule between DNA expression and protein translation. Because of its unique properties (such as its typically single-stranded nature and its 2' OH group) and its ability to adopt many different secondary/tertiary structures, both coding and noncoding RNAs have attracted attention in medicine. Research has begun to explore RNAs potential to be used for therapeutic benefit, and unique challenges have occurred during drug discovery and implementation of RNA therapeutics.[10]
mRNA
Messenger RNA (mRNA) is a single-stranded RNA molecule that is complementary to one of the DNA strands of a gene.[11] An mRNA molecule transfers a portion of the DNA code to other parts of the cell for making proteins.[12] DNA therapeutics needs access to the nucleus to be transcribed into RNA, and its functionality depends on nuclear envelope breakdown during cell division. However, mRNA therapeutics do not need to enter into the nucleus to be functional since it will be translated immediately once it has reached to the cytoplasm.[13] Moreover, unlike plasmids and viral vectors, mRNAs do not integrate into the genome and therefore do not have the risk of insertional mutagenesis,[14] making them suitable for use in cancer vaccines, tumor immunotherapy and infectious disease prevention.[15]
Discovery and development
In 1953, Alfred Day Hershey reported that soon after infection with phage, bacteria produced a form of RNA at a high level and this RNA was also broken down rapidly.[16] However, the first clear indication of mRNA was from the work of Elliot Volkin and Lazarus Astrachan in 1956 by infecting E.coli with T2 bacteriophages and putting them into the medium with 32P.[17][18] They found out that the protein synthesis of E.coli was stopped and phage proteins were synthesized.[19] Then, in May 1961, their collaborated researchers Sydney Brenner, François Jacob, and Jim Watson announced the isolation of mRNA.[20][21] For a few decades after mRNA discovery, people focused on understanding the structural, functional, and metabolism pathway aspects of mRNAs. However, in 1990, Jon A. Wolff demonstrated the idea of nucleic acid-encoded drugs by direct injecting in vitro transcribed (IVT) mRNA or plasmid DNA (pDNA) into the skeletal muscle of mice which expressed the encoded protein in the injected muscle.[22][23][24]
Once IVT mRNA has reached the cytoplasm, the mRNA is translated instantly. Thus, it does not need to enter the nucleus to be functional.[25] Also, it does not integrate into the genome and therefore does not have the risk of insertional mutagenesis.[26] Moreover, IVT mRNA is only transiently active and is completely degraded via physiological metabolic pathways.[27] Due to these reasons, IVT mRNA has undergone extensive preclinical investigation.
Mechanisms
In vitro transcription (IVT) is performed on a linearized DNA plasmid template containing the targeted coding sequence. Then, naked mRNA or mRNA complexed in a nanoparticle will be delivered systemically or locally. Subsequently, a part of the exogenous naked mRNA or complexed mRNA will go through cell-specific mechanisms. Once in the cytoplasm, the IVT mRNA is translated by the protein synthesis machinery.[28][29]
There are two identified RNA sensors, toll-like receptors (TLRs) and the RIG-I-like receptor family. TLRs are localized in the endosomal compartment of cells, such as DCs and macrophages.[30] RIG-I-like family is as a pattern recognition receptor (PRR).[31] However, the immune response mechanisms and process of mRNA vaccine recognition by cellular sensors and the mechanism of sensor activation are still unclear.[29]
Cancer immunotherapy
In 1995, Robert Conry demonstrated that intramuscular injection of naked RNA encoding carcinoembryonic antigen elicited antigen-specific antibody responses.[32] Then, it was elaborated by demonstrating that dendritic cells(DCs) exposed to mRNA coding for specific antigens or to total mRNA extracted from tumor cells and injected into tumor-bearing mice induced T cell immune responses and inhibited the growth of tumors.[33] Then, researchers started to approach mRNA transfected DCs using vaccines based on ex vivo IVT mRNA-transfected DCs.[34] Meanwhile, Argos Therapeutics had initiated a Phase III clinical trial using DCs with advanced renal cell carcinoma in 2015 (NCT01582672) but it was terminated due to the lack of efficacy.[35]
For further application, IVT mRNA was optimized for in situ transfections of DCs in vivo. It improved the translation efficiency and stability of IVT mRNA and enhanced the presentation of the mRNA-encoded antigen on MHC class I and II molecules.[36][37] Then, they found out that the direct injection of naked IVT mRNA into lymph nodes was the most effective way to induce T cell responses.[38] Based on this discovery, first-in-human testing of the injection of naked IVT mRNA encoding cancer antigens by BioNTech has started with patients with melanoma (NCT01684241).[39]
Recently, the new cancer immunotherapy, the combining of self-delivering RNA(sd-rxRNA) and adoptive cell transfer(ACT) therapy, was invented by RXi Pharmaceuticals and the Karolinska Institute. In this therapy, the sd-rxRNA eliminated the expression of immunosuppressive receptors and proteins in therapeutic immune cells so it improved the ability of immune cells to destroy the tumor cells. Then, the PD-1 targeted sd-rxRNA helped increasing the anti-tumor activity of tumor-infiltrating lymphocytes (TIL) against melanoma cells.[40][41] Based on this idea, the mRNA-4157 has been tested and passed phase I clinical trial.[42]
Cytosolic nucleic acid-sensing pathways can enhance immune response to cancer. RIG-I agonist, stem loop RNA (SLR) 14. Tumor growth was significantly delayed and extended survival in mice. SLR14 improved antitumor efficacy of anti-PD1 antibody over single-agent treatment. SLR14 was absorbed by CD11b+ myeloid cells in the tumor microenvironment. Genes associated with immune defense were significantly up-regulated, along with increased CD8+ T lymphocytes, NK cells, and CD11b+ cells. SLR14 inhibited nonimmunogenic B16 tumor growth, leaving immune memory.[43]
Vaccines
In 1993, the first success of an mRNA vaccine was reported in mice, by using liposome-encapsulated IVT mRNA which is encoding the nucleoprotein of influenza that induced virus-specific T cells.[44] Then, IVT mRNA was formulated with synthetic lipid nanoparticles and it induced protective antibody responses against the respiratory syncytial virus(RSV) and influenza virus in mice.[45]
There are a few different types of IVT mRNA-based vaccine development for infectious diseases. One of the successful types is using self-amplifying IVT mRNA that has sequences of positive-stranded RNA viruses. It was originally developed for a flavivirus and it was workable with intradermal injection. One of the other ways is injecting a two-component vaccine which is containing an mRNA adjuvant and naked IVT mRNA encoding influenza hemagglutinin antigen only or in combination with neuraminidase encoding IVT mRNA.[46]
For example, for the HIV treatment, vaccines are using DCs transfected with IVT mRNA that is encoding HIV proteins. There are a few phase I and II clinical trials using IVT mRNA encoding combinations and it shows that antigen-specific CD8+ and CD4+ T cell responses can be induced. However, no antiviral effects have been observed in the clinical trial.[47][48]
One of the other mRNA vaccines is for COVID-19. The Severe Acute Respiratory Syndrome CoronaVirus 2 (SARS-CoV-2) outbreaks in December 2019 and spread all over the world, causing a pandemic of respiratory illness designated coronavirus disease 2019 (COVID-19).[49] The Moderna COVID-19 vaccine, manufactured by Moderna since 2020, is a lipid nanoparticle (LNP) encapsulated mRNA-based vaccine that encodes for a full-length, prefusion stabilized spike(S)-2P antigen of SARS-CoV-2 with a transmembrane anchor.[50][51]
Anti-viral
In 2021, SLR14 was reported to prevent infection in the lower respiratory tract and severe disease in an interferon type I (IFN-I)–dependent manner in mice. Immunodeficient mice with chronic SARS-CoV-2 infection experienced near-sterilizing innate immunity with no help from the adaptive immune system.[52]
Tissue regeneration
A 2022 study by researchers from the Mayo Clinic, Maastricht University, and Ethris GmBH, a biotech company that focuses on RNA therapeutics, found that chemically modified mRNA encoding BMP-2 promoted dosage-dependent healing of femoral osteotomies in male rats. The mRNA molecules were complexed within nonviral lipid particles, loaded onto sponges, and surgically implanted into the bone defects. They remained localized around the site of application. Compared to receiving rhBMP-2 directly, bony tissues regenerated after mRNA treatment displayed superior strength and less formation of massive callus.[53]
Limitations
There are many challenges for the successful translation of mRNA into drugs because mRNA is a very large and heavy molecule(10^5 ~ 10^6 Da). Moreover, mRNA is unstable and easily degraded by nucleases, and it also activates the immune systems.[54] Furthermore, mRNA has a high negative charge density and it reduces the permeation of mRNA across cellular membranes.[55] Due to these reasons, without the appropriate delivery system, mRNA is degraded easily and the half-life of mRNA without a delivery system is only around 7 hours.[56] Even though some degrees of challenges could be overcome by chemical modifications, delivery of mRNA remains an obstacle. The methods that have been researched to improve the delivery system of mRNA are using microinjection, RNA patches (mRNA loaded in a dissolving micro-needle), gene gun, protamine condensation, RNA adjuvants, and encapsulating mRNA in nanoparticles with lipids.[54][57][58]
Even though In Vitro Translated (IVT) mRNA with delivery agents showed improved resistance against degradation, it needs more studies on how to improve the efficiency of the delivery of naked mRNA in vivo.[23]
Antisense RNA
Antisense RNA is the non-coding and single-stranded RNA that is complementary to a coding sequence of mRNA. It inhibits the ability of mRNA to be translated into proteins.[59] Short antisense RNA transcripts are produced within the nucleus by the action of the enzyme Dicer, which cleaves double-stranded RNA precursors into 21–26 nucleotide long RNA species.[4]
There is an antisense-based discovery strategy, rationale and design of screening assays, and the application of such assays for screening of natural product extracts and the discovery of fatty acid condensing enzyme inhibitors.[60] Antisense RNA is used for treating cancer and inhibition of metastasis and vectors for antisense sequestration. Particularly MicroRNAs(miRs) 15 and 16 to a patient in need of the treatment for diagnosis and prophylaxis of cancer.[61] Antisense drugs are based on the fact that antisense RNA hybridizes with and inactivates mRNA. These drugs are short sequences of RNA that attach to mRNA and stop a particular gene from producing the protein for which it encodes. Antisense drugs are being developed to treat lung cancer, diabetes and diseases such as arthritis and asthma with a major inflammatory component.[62] It shows that the decreased expression of MLLT4 antisense RNA 1 (MLLT4‑AS1) is a potential biomarker and a predictor of a poor prognosis for gastric cancer. So far, applications of antisense RNAs in antivirus and anticancer treatments and in regulating the expression of related genes in plants and microorganisms have been explored.[63]
Non-viral vectors, virus vectors and liposomes have been used to deliver the antisense RNA through the cell membrane into the cytoplasm and nucleus. It has been found that the viral vector based delivery is the most advantageous among different delivery systems because it has a high transfection efficacy.[64] However, it is difficult to deliver antisense RNA only to the targeted sites. Also, due to the size and the stability issues of antisense RNA, there are some limitations to its use. To improve the delivery issues, chemical modifications, and new oligonucleotide designs have been studied to enhance the drug distribution, side effects, and tolerability.[65][66]
RNAi
Interfering RNA are a class of short, noncoding RNA that act to translationally or post-translationally repress gene expression.[67][5] Their discovery and subsequent identification as key effectors of post-transcriptional gene regulation have made small interfering RNA (siRNA) and micro RNA (miRNA) potential therapeutics for systemic diseases.[67][5][68] The RNAi system was originally discovered in 1990 by Jorgensen et al., who were doing research involving the introduction of coloration genes into petunias,[68][69] and it is thought that this system originally developed as a means of innate immunity against double-stranded RNA viruses.[70]
siRNA
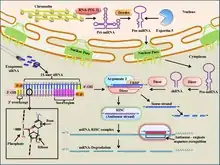
Small interfering (siRNA) are short, 19-23 base-pair (with a 3' overhang of two nucleotides), double-stranded pieces of RNA that participate in the RNA-induced silencing complex (RISC) for gene silencing.[5][68] Specifically, siRNA is bound by the RISC complex where it is unwound using ATP hydrolysis.[68][71][72] It is then used as a guide by the enzyme "Slicer" to target mRNAs for degradation based on complementary base-pairing to the target mRNA.[68][71][72] As a therapeutic, siRNA is able to be delivered locally, through the eye or nose, to treat various diseases.[5] Local delivery benefits from simple formulation and drug delivery and high bioavailability of the drug.[5] Systemic delivery is necessary to target cancers and other diseases.[5] Targeting the siRNA when delivered locally is one of the main challenges in siRNA therapeutics.[5] While it is possible to use intravenous injection to deliver siRNA therapies, concerns have been raised about the large volumes used in the injection, as these must often be ~20-30% of the total blood volume.[68] Other methods of delivery include liposome packaging, conjugation to membrane-permeable peptides, and direct tissue/organ electroporation.[68] Additionally, it has been found that exogeneous siRNAs only last a few days (a few weeks at most in non-dividing cells) in vivo.[73][74] If siRNA is able to successfully reach its target, it has the potential to therapeutically regulate gene expression through its ability to base-pair to mRNA targets and promote their degradation through the RISC system[5][68] Currently, siRNA-based therapy is in a phase I clinical trial for the treatment of age-related macular degeneration,[68] although it is also being explored for use in cancer therapy. For instance, siRNA can be used to target mRNAs that code for proteins that promote tumor growth such as the VEGF receptor and telomerase enzyme.[68]
miRNA
Micro RNAs (miRNAs) are short, ~19-23 base pair long RNA oligonucleotides that are involved in the microRNA-induced silencing complex.[68][6] Specifically, once loaded onto the ARGONAUTE enzyme, miRNAs work with mRNAs to repress translation and post-translationally destabilize mRNA.[6] While they are functionally similar to siRNAs, miRNAs do not require extensive base-pairing for mRNA silencing (can require as few as seven base-pairs with target),[75][76] thus allowing them to broadly affect a wider range of mRNA targets.[77] In the cell, miRNA uses switch, tuning, and neutral interactions to finely regulate gene repression.[6] As a therapeutic, miRNA has the potential to affect biochemical pathways throughout the organism.[67]
With more than 400 miRNA identified in humans, discerning their target gene for repression is the first challenge.[6] Multiple databases have been built, for example TargetScan, using miRNA seed matching.[67] In vitro assays assist in determining the phenotypic effects of miRNAs,[67] but due to the complex nature of gene regulation not all identified miRNAs have the expected effect.[6] Additionally, several miRNAs have been found to act as either tumor suppressors or oncogenes in vivo, such as the oncogenic miR-155 and miR-17-92.[77]
In clinical trials, miRNA are commonly used as biomarkers for a variety of diseases, potentially providing earlier diagnosis as well as disease progression, stage, and genetic links.[67] Phase 1 and 2 trials currently test miRNA mimics (to express genes) and miRNA (to repress genes) in patients with cancers and other diseases.[67] In particular, mimic miRNAs are used to introduce miRNAs that act as tumor suppressors into cancerous tissues, while miRNA antagonists are used to target oncogenic miRNAs to prevent their cancer-promoting activity.[77] Therapeutic miRNA is also used in addition to common therapies (such as cancer therapies) that are known to overexpress or destabilize the patient miRNA levels.[67] An example of one mimic miRNA therapy that demonstrated efficacy in impeding lung cancer tumor growth in mouse studies is miR-34a.[77][78]
One concerning aspect of miRNA-based therapies is the potential for the exogeneous miRNA to affect miRNA silencing mechanisms within normal body cells, thereby affecting normal cellular biochemical pathways.[77] However, in vivo studies have indicated that miRNAs display little to no effect in non-target tissues/organs.[78][79]
RNA aptamers
Broadly, aptamers are small molecules composed of either single-stranded DNA or RNA and are typically 20-100 nucleotides in length,[7][8][81] or ~3-60 kDa.[81][82] Because of their single-stranded nature, aptamers are capable of forming many secondary structures, including pseudoknots, stem loops, and bulges, through intra-strand base pairing interactions.[81] The combinations of secondary structures present in an aptamer confer it a particular tertiary structure which in turn dictates the specific target the aptamer will selectively bind to.[81][83] Because of the selective binding ability of aptamers, they are considering a promising biomolecule for use in pharmaceuticals.[7][8][81] Additionally, aptamers exhibit tight binding to targets, with dissociation constants often in the pM to nM range.[7][9] Besides their strong binding ability, aptamers are also valued because they can be used on targets that are not capable of being bound by small peptides generated by phage display or by antibodies, and they are able to differentiate between conformational isomers and amino acid substitutions.[81][84][85] Also, because aptamers are nucleic-acid based, they can be directly synthesized, eliminating the need for cell-based expression and extraction as is the case in antibody production.[8][86] RNA aptamers in particular are capable of producing a myriad of different structures, leading to speculations that they are more discriminating in their target affinity compared to DNA aptamers.[81][87]
Discovery and development
Aptamers were originally discovered in 1990 when Lary Gold and Craig Tuerk utilized a method of directed evolution known as SELEX to isolate a small single stranded RNA molecule that was capable of binding to T4 bacteriophage DNA polymerase.[8][88] Additionally, the term “aptamer” was coined by Andrew Ellington, who worked with Jack Szostak to select an RNA aptamer that was capable of tight binding to certain organic dye molecules.[81][89] The term itself is a conglomeration of the Latin “aptus” or “to fit” and the Greek “meros” or “part."[81][89]
RNA aptamers are not so much “created” as “selected.” To develop an RNA aptamer capable of selective binding to a molecular target, a method known as Systematic Evolution of Ligands by EXponential Enrichment (SELEX) is used to isolate a unique RNA aptamer from a pool of ~10^13 to 10^16 different aptamers, otherwise known as a library.[7][8][81][88][89] The library of potential aptamer oligonucleotides is then incubated with a non-target species so as to remove aptamers that exhibit non-specific binding.[7] After subsequent removal of the non-specific aptamers, the remaining library members are then exposed to the desired target, which can be a protein, peptide, cell type, or even an organ (in the case of live animal-based SELEX).[7][81][90][91][92][93][94][95] From there, the RNA aptamers which were bound to the target are transcribed to cDNA which then is amplified through PCR, and the PCR products are then re-transcribed to RNA.[81] These new RNA transcripts are then used to repeat the selection cycle many times, thus eventually producing a homogeneous pool of RNA aptamers capable of highly specific, high-affinity target binding.[7]
Examples
RNA aptamers can be designed to act as antagonists, agonists, or so-called ”RNA decoy aptamers."[81][96] In the case of antagonists, the RNA aptamer is used either to prevent binding of a certain protein to its cell membrane receptor or to prevent the protein from performing its activity by binding to the protein's target.[81] Currently, the only RNA aptamer-based therapies that have advanced to clinical trials act as antagonists.[81] When RNA aptamers are designed to act as agonists, they promote immune cell activation as a co-stimulatory molecule, thus aiding in the mobilization of the body's own defense system.[81][97] For RNA decoy aptamers, the synthetic RNA aptamer resembles a native RNA molecule.[81][96] As such, proteins(s) which bind to the native RNA target instead bind to the RNA aptamer, possibly interfering with the biomolecular pathway of a particular disease.[81][96] In addition to their utility as direct therapeutic agents, RNA aptamers are also being considered for other therapeutic roles. For instance, by conjugating the RNA aptamer to a drug compound, the RNA aptamer can act as a targeted delivery system for that drug.[7] Such RNA aptamers are known as ApDCs.[7] Additionally, through conjugation to radioisotope or a fluorescent dye molecule, RNA aptamers may be useful in diagnostic imaging.[7][98][99]
Because of the SELEX process utilized to select RNA aptamers, RNA aptamers can be generated for many potential targets. By directly introducing the RNA aptamers to the target during SELEX, a very selective, high-affinity, homogeneous pool of RNA aptamers can be produced. As such, RNA aptamers can be made to target small peptides and proteins, as well as cell fragments, whole cells, and even specific tissues.[7][81][100][101][93][102] Examples of RNA aptamer molecular targets and potential targets include vascular endothelial growth factor,[103] osteoblasts,[104] and C-X-C Chemokine Ligand 12 (CXCL2).[7][8][105]
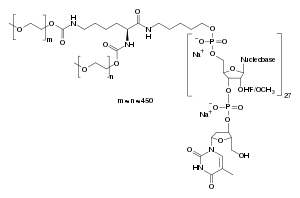
An example of an RNA aptamer therapy includes Pegaptanib (aka Macugen ® ), the only FDA-approved RNA aptamer treatment.[7][8][81] Originally approved in 2004 to treat age-related macular degeneration, Pegaptanib is a 28 nucleotide RNA aptamer that acts as a VEGF antagonist.[7][8][81] However, it is not as effective as antibody-based treatments such as bevacizumab and ranibizumab.[81][106][107] Another example of an RNA aptamer therapeutic is NOX-A12, a 45 nucleotide RNA aptamer that is in clinical trials for chronic lymphocytic leukemia, pancreatic cancer, as well as other cancers.[8] NOX-A12 acts as antagonist for CXCL12/SDF-1, a chemokine involved in tumor growth.[8]
Limitations
While the high-selectivity and tight-binding of RNA aptamers have generated interest in their use as pharmaceuticals, there are many problems which have prevented them from being successful in vivo. For one, without modifications RNA aptamers are degraded after being introduced into the body by nucleases in the span of a few minutes.[8][81][108][109] Also, due to their small size, RNA aptamers can be removed from the bloodstream by the renal system.[8][81][82][108][109] Because of their negative charge, RNA aptamers are additionally known to bind proteins in the bloodstream, leading to non-target tissue delivery and toxicity.[81][110][111] Care must also be taken when isolating the RNA aptamers, as aptamers which contain repeated Cytosine-Phosphate-Guanine (CpG) sequences will cause immune system activation through the Toll-like receptor pathway.[8][112][113]
In order to combat some of the in vivo limitations of RNA aptamers, various modifications can be added to the nucleotides to aid in efficacy of the aptamer. For instance, a polyethylene glycol (PEG) moiety can be attached to increase the size of the aptamer, thereby preventing its removal from the bloodstream by the renal glomerulus.[114][115] However, PEG has been implicated in allergic reactions during in vivo testing.[81][116][117] Furthermore, modifications can be added to prevent nuclease degradation, such as a 2’ fluoro or amino group as well as a 3’ inverted thymidine.[8][81][118][119] Additionally, the aptamer can be synthesized so that the ribose sugar is in the L-form instead of the D-form, further preventing nuclease recognition.[7][81][120][121] Such aptamers are known as Spiegelmers.[7][121] In order to prevent Toll-like receptor pathway activation, the cytosine nucleobases within the aptamer can be methylated.[8] Nevertheless, despite these potential solutions to reduced in vivo efficacy, it is possible that chemically modifying the aptamer may weaken its binding affinity towards its target.[81][122]
See also
- Riboswitch
- ncRNA therapy
References
- Sahin, U.; Karikó, K.; Türeci, Ö. (2014), "mRNA-based therapeutics – developing a new class of drugs", Nature Reviews. Drug Discovery, 13 (10): 759–780, doi:10.1038/nrd4278, PMID 25233993, S2CID 27454546
- "Our COVID-19 Vaccine Study – What's Next? | Pfizer". www.pfizer.com. Retrieved 2020-11-29.
- DeFrancesco L (March 2017). "The 'anti-hype' vaccine". Nature Biotechnology. 35 (3): 193–197. doi:10.1038/nbt.3812. PMID 28244993. S2CID 3606308.
- Iida T, Nakayama J, Moazed D (July 2008). "siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription". Molecular Cell. 31 (2): 178–89. doi:10.1016/j.molcel.2008.07.003. PMC 2575423. PMID 18657501.
- Nikam RR, Gore KR (August 2018). "Journey of siRNA: Clinical Developments and Targeted Delivery". Nucleic Acid Therapeutics. 28 (4): 209–224. doi:10.1089/nat.2017.0715. PMID 29584585. S2CID 4763853.
- Bartel DP (January 2009). "MicroRNAs: target recognition and regulatory functions". Cell. 136 (2): 215–33. doi:10.1016/j.cell.2009.01.002. PMC 3794896. PMID 19167326.
- Zhu G, Chen X (September 2018). "Aptamer-based targeted therapy". Advanced Drug Delivery Reviews. 134: 65–78. doi:10.1016/j.addr.2018.08.005. PMC 6239901. PMID 30125604.
- Kaur H, Bruno JG, Kumar A, Sharma TK (2018-07-01). "Aptamers in the Therapeutics and Diagnostics Pipelines". Theranostics. 8 (15): 4016–4032. doi:10.7150/thno.25958. PMC 6096388. PMID 30128033.
- Dunn MR, Jimenez RM, Chaput JC (2017-10-04). "Analysis of aptamer discovery and technology". Nature Reviews Chemistry. 1 (10): 1–16. doi:10.1038/s41570-017-0076. ISSN 2397-3358.
- Coutinho MF, Matos L, Santos JI, Alves S (2019). Romão L (ed.). The mRNA Metabolism in Human Disease. Advances in Experimental Medicine and Biology. Vol. 1157. Cham: Springer International Publishing. pp. 133–177. doi:10.1007/978-3-030-19966-1_7. ISBN 978-3-030-19965-4. PMID 31342441. S2CID 198491788.
- "Messenger RNA (mRNA)". Genome.gov. Retrieved 2020-10-05.
- "mRNA". Biology Dictionary. 2016-11-25. Retrieved 2020-11-30.
- Herndon MK, Quirk CC, Nilson JH (January 2016). "Chapter 2 - Control of Hormone Gene Expression". In Jameson JL, De Groot LJ, de Kretser DM, Giudice LC (eds.). Endocrinology: Adult and Pediatric (Seventh ed.). Philadelphia: W.B. Saunders. pp. 16–29.e2. doi:10.1016/B978-0-323-18907-1.00002-0. ISBN 978-0-323-18907-1.
- Yamamoto A, Kormann M, Rosenecker J, Rudolph C (March 2009). "Current prospects for mRNA gene delivery". European Journal of Pharmaceutics and Biopharmaceutics. 71 (3): 484–9. doi:10.1016/j.ejpb.2008.09.016. PMID 18948192.
- Huang L, Zhang L, Li W, Li S, Wen J, Li H, Liu Z (July 2020). "Advances in Development of mRNA-Based Therapeutics". Current Topics in Microbiology and Immunology. Berlin, Heidelberg: Springer: 1–20. doi:10.1007/82_2020_222. PMID 32683507. S2CID 220654211.
- Hershey AD (September 1953). "Nucleic acid economy in bacteria infected with bacteriophage T2". The Journal of General Physiology. 37 (1): 1–23. doi:10.1085/jgp.37.1.1. PMC 2147426. PMID 13084888.
- "Elliot Volkin Obituary (2012) Knoxville News Sentinel". Legacy.com. Retrieved 2021-07-01.
- Wade, Nicholas (2003-08-02). "Lazarus Astrachan, 78, Conducted RNA Studies". The New York Times. ISSN 0362-4331. Retrieved 2021-07-01.
- Astrachan L, Colowick CP, Kaplan NO (April 1956). "Phosphorus incorporation in Escherichia coli ribonucleic acid after infection with bacteriophage T2". Virology. 2 (2): 149–161. doi:10.1016/0042-6822(56)90016-2. PMID 13312220.
- Brenner S, Jacob F, Meselson M (May 1961). "An unstable intermediate carrying information from genes to ribosomes for protein synthesis". Nature. 190 (4776): 576–581. Bibcode:1961Natur.190..576B. doi:10.1038/190576a0. PMID 20446365. S2CID 4200865.
- Gros F, Hiatt H, Gilbert W, Kurland CG, Risebrough RW, Watson JD (May 1961). "Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli". Nature. 190 (4776): 581–5. Bibcode:1961Natur.190..581G. doi:10.1038/190581a0. PMID 13708983. S2CID 4197056.
- Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL (March 1990). "Direct gene transfer into mouse muscle in vivo". Science. 247 (4949 Pt 1): 1465–8. Bibcode:1990Sci...247.1465W. doi:10.1126/science.1690918. PMID 1690918.
- Sahin U, Karikó K, Türeci Ö (October 2014). "mRNA-based therapeutics--developing a new class of drugs". Nature Reviews. Drug Discovery. 13 (10): 759–80. doi:10.1038/nrd4278. PMID 25233993. S2CID 27454546.
- Beckert, Bertrand; Masquida, Benoît (2011). "Synthesis of RNA by in vitro transcription". RNA. Methods in Molecular Biology (Clifton, N.J.). Vol. 703. pp. 29–41. doi:10.1007/978-1-59745-248-9_3. ISBN 978-1-58829-913-0. ISSN 1940-6029. PMID 21125481.
- Karikó K (April 2019). "In vitro-Transcribed mRNA Therapeutics: Out of the Shadows and Into the Spotlight". Molecular Therapy. 27 (4): 691–692. doi:10.1016/j.ymthe.2019.03.009. PMC 6453554. PMID 30905578.
- Mandl CW, Aberle JH, Aberle SW, Holzmann H, Allison SL, Heinz FX (December 1998). "In vitro-synthesized infectious RNA as an attenuated live vaccine in a flavivirus model". Nature Medicine. 4 (12): 1438–40. doi:10.1038/4031. PMID 9846585. S2CID 22676794.
- Hoerr I, Obst R, Rammensee HG, Jung G (January 2000). "In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies". European Journal of Immunology. 30 (1): 1–7. doi:10.1002/1521-4141(200001)30:1<1::aid-immu1>3.0.co;2-#. PMID 10602021. S2CID 196606232.
- Van Hoecke L, Roose K (February 2019). "How mRNA therapeutics are entering the monoclonal antibody field". Journal of Translational Medicine. 17 (1): 54. doi:10.1186/s12967-019-1804-8. PMC 6387507. PMID 30795778.
- Zhang C, Maruggi G, Shan H, Li J (2019). "Advances in mRNA Vaccines for Infectious Diseases". Frontiers in Immunology. 10: 594. doi:10.3389/fimmu.2019.00594. PMC 6446947. PMID 30972078.
- Hua Z, Hou B (March 2013). "TLR signaling in B-cell development and activation". Cellular & Molecular Immunology. 10 (2): 103–6. doi:10.1038/cmi.2012.61. PMC 4003046. PMID 23241902.
- Kato H, Oh SW, Fujita T (May 2017). "RIG-I-Like Receptors and Type I Interferonopathies". Journal of Interferon & Cytokine Research. 37 (5): 207–213. doi:10.1089/jir.2016.0095. PMC 5439449. PMID 28475461.
- Conry RM, LoBuglio AF, Wright M, Sumerel L, Pike MJ, Johanning F, et al. (April 1995). "Characterization of a messenger RNA polynucleotide vaccine vector". Cancer Research. 55 (7): 1397–400. PMID 7882341.
- Boczkowski D, Nair SK, Snyder D, Gilboa E (August 1996). "Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo". The Journal of Experimental Medicine. 184 (2): 465–72. doi:10.1084/jem.184.2.465. PMC 2192710. PMID 8760800.
- Nair SK, Heiser A, Boczkowski D, Majumdar A, Naoe M, Lebkowski JS, et al. (September 2000). "Induction of cytotoxic T cell responses and tumor immunity against unrelated tumors using telomerase reverse transcriptase RNA transfected dendritic cells". Nature Medicine. 6 (9): 1011–7. doi:10.1038/79519. PMID 10973321. S2CID 6109427.
- Argos Therapeutics (2018-06-13). "An International Phase 3 Randomized Trial of Autologous Dendritic Cell Immunotherapy (AGS-003) Plus Standard Treatment of Advanced Renal Cell Carcinoma (ADAPT)". ClinicalTrials. Retrieved 30 November 2020.
- Kreiter S, Konrad T, Sester M, Huber C, Türeci O, Sahin U (October 2007). "Simultaneous ex vivo quantification of antigen-specific CD4+ and CD8+ T cell responses using in vitro transcribed RNA". Cancer Immunology, Immunotherapy. 56 (10): 1577–87. doi:10.1007/s00262-007-0302-7. PMID 17361438. S2CID 23374448.
- Kreiter S, Selmi A, Diken M, Sebastian M, Osterloh P, Schild H, et al. (January 2008). "Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals". Journal of Immunology. 180 (1): 309–18. doi:10.4049/jimmunol.180.1.309. PMID 18097032. S2CID 83698413.
- Kuhn AN, Diken M, Kreiter S, Selmi A, Kowalska J, Jemielity J, et al. (August 2010). "Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo". Gene Therapy. 17 (8): 961–71. doi:10.1038/gt.2010.52. PMID 20410931. S2CID 647837.
- BioNTech RNA Pharmaceuticals GmbH (2020-01-14). "Clinical First-in-human Dose Escalation Study Evaluating the Safety and Tolerability of Intranodal Administration of an RNA-based Cancer Vaccine Targeting Two Tumor-associated Antigens in Patients With Advanced Melanoma". ClinicalTrials. Retrieved 30 November 2020.
- Ligtenberg MA, Pico de Coaña Y, Shmushkovich T, Yoshimoto Y, Truxova I, Yang Y, et al. (June 2018). "Self-Delivering RNAi Targeting PD-1 Improves Tumor-Specific T Cell Functionality for Adoptive Cell Therapy of Malignant Melanoma". Molecular Therapy. 26 (6): 1482–1493. doi:10.1016/j.ymthe.2018.04.015. PMC 5986970. PMID 29735366.
- Lian S, Xie R, Ye Y, Xie X, Li S, Lu Y, et al. (April 2019). "Simultaneous blocking of CD47 and PD-L1 increases innate and adaptive cancer immune responses and cytokine release". EBioMedicine. 42: 281–295. doi:10.1016/j.ebiom.2019.03.018. PMC 6491392. PMID 30878596.
- "mRNA-4157 | Moderna, Inc". www.modernatx.com. Retrieved 2020-11-30.
- Jiang, Xiaodong; Muthusamy, Viswanathan; Fedorova, Olga; Kong, Yong; Kim, Daniel J.; Bosenberg, Marcus; Pyle, Anna Marie; Iwasaki, Akiko (2019-12-02). "Intratumoral delivery of RIG-I agonist SLR14 induces robust antitumor responses". The Journal of Experimental Medicine. 216 (12): 2854–2868. doi:10.1084/jem.20190801. ISSN 1540-9538. PMC 6888973. PMID 31601678.
- Martinon F, Krishnan S, Lenzen G, Magné R, Gomard E, Guillet JG, et al. (July 1993). "Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA". European Journal of Immunology. 23 (7): 1719–22. doi:10.1002/eji.1830230749. PMID 8325342. S2CID 42640967.
- Hekele A, Bertholet S, Archer J, Gibson DG, Palladino G, Brito LA, et al. (August 2013). "Rapidly produced SAM(®) vaccine against H7N9 influenza is immunogenic in mice". Emerging Microbes & Infections. 2 (8): e52. doi:10.1038/emi.2013.54. PMC 3821287. PMID 26038486.
- Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, et al. (December 2012). "Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection". Nature Biotechnology. 30 (12): 1210–6. doi:10.1038/nbt.2436. PMID 23159882. S2CID 12488462.
- Routy JP, Boulassel MR, Yassine-Diab B, Nicolette C, Healey D, Jain R, et al. (February 2010). "Immunologic activity and safety of autologous HIV RNA-electroporated dendritic cells in HIV-1 infected patients receiving antiretroviral therapy". Clinical Immunology. 134 (2): 140–7. doi:10.1016/j.clim.2009.09.009. PMC 2818410. PMID 19889582.
- Van Gulck E, Vlieghe E, Vekemans M, Van Tendeloo VF, Van De Velde A, Smits E, et al. (February 2012). "mRNA-based dendritic cell vaccination induces potent antiviral T-cell responses in HIV-1-infected patients". AIDS. 26 (4): F1-12. doi:10.1097/qad.0b013e32834f33e8. PMID 22156965. S2CID 205988884.
- Helmy YA, Fawzy M, Elaswad A, Sobieh A, Kenney SP, Shehata AA (April 2020). "The COVID-19 Pandemic: A Comprehensive Review of Taxonomy, Genetics, Epidemiology, Diagnosis, Treatment, and Control". Journal of Clinical Medicine. 9 (4): 1225. doi:10.3390/jcm9041225. PMC 7230578. PMID 32344679.
- "Drug". Default. Retrieved 2020-11-30.
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. (November 2020). "An mRNA Vaccine against SARS-CoV-2 - Preliminary Report". The New England Journal of Medicine. 383 (20): 1920–1931. doi:10.1056/NEJMoa2022483. PMC 7377258. PMID 32663912.
- Mao, Tianyang; Israelow, Benjamin; Lucas, Carolina; Vogels, Chantal B.F.; Gomez-Calvo, Maria Luisa; Fedorova, Olga; Breban, Mallery I.; Menasche, Bridget L.; Dong, Huiping; Linehan, Melissa; Yale SARS-CoV-2 Genome Surveillance Initiative (2021-11-10). "A stem-loop RNA RIG-I agonist protects against acute and chronic SARS-CoV-2 infection in mice". Journal of Experimental Medicine. 219 (1): e20211818. doi:10.1084/jem.20211818. ISSN 0022-1007. PMC 8590200. PMID 34757384.
- De La Vega, Rodolfo E.; van Griensven, Martijn; Zhang, Wen; Coenen, Michael J.; Nagelli, Christopher V.; Panos, Joseph A.; Peniche Silva, Carlos J.; Geiger, Johannes; Plank, Christian; Evans, Christopher H.; Balmayor, Elizabeth R. (2022-02-18). "Efficient healing of large osseous segmental defects using optimized chemically modified messenger RNA encoding BMP-2". Science Advances. 8 (7): eabl6242. Bibcode:2022SciA....8.6242D. doi:10.1126/sciadv.abl6242. ISSN 2375-2548. PMC 8849297. PMID 35171668.
- Houseley J, Tollervey D (February 2009). "The many pathways of RNA degradation". Cell. 136 (4): 763–76. doi:10.1016/j.cell.2009.01.019. PMID 19239894. S2CID 17570967.
- Kowalski PS, Rudra A, Miao L, Anderson DG (April 2019). "Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery". Molecular Therapy. 27 (4): 710–728. doi:10.1016/j.ymthe.2019.02.012. PMC 6453548. PMID 30846391.
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS (February 2009). "Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells". DNA Research. 16 (1): 45–58. doi:10.1093/dnares/dsn030. PMC 2644350. PMID 19001483.
- Deering RP, Kommareddy S, Ulmer JB, Brito LA, Geall AJ (June 2014). "Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines". Expert Opinion on Drug Delivery. 11 (6): 885–99. doi:10.1517/17425247.2014.901308. PMID 24665982. S2CID 33489182.
- Golombek S, Pilz M, Steinle H, Kochba E, Levin Y, Lunter D, et al. (June 2018). "Intradermal Delivery of Synthetic mRNA Using Hollow Microneedles for Efficient and Rapid Production of Exogenous Proteins in Skin". Molecular Therapy: Nucleic Acids. 11: 382–392. doi:10.1016/j.omtn.2018.03.005. PMC 5992458. PMID 29858073.
- "7.19A: RNA Regulation and Antisense RNA". Biology LibreTexts. 2017-06-06. Retrieved 2020-12-01.
- Singh SB, Phillips JW, Wang J (March 2007). "Highly sensitive target-based whole-cell antibacterial discovery strategy by antisense RNA silencing". Current Opinion in Drug Discovery & Development. 10 (2): 160–6. PMID 17436551.
- US 2017283805, Bonci D, De Maria R, "Antisense RNA for Treating Cancer and Inhibition of Metastasis and Vectors for Antisense Sequestration", published 2017, assigned to Istituto Superiore di Sanità
- Wade JT (2013). "Antisense RNA". Brenner's Encyclopedia of Genetics. doi:10.1016/B978-0-12-809633-8.06068-4. ISBN 978-0-12-809633-8.
- Xu JZ, Zhang JL, Zhang WG (2018). "Antisense RNA: the new favorite in genetic research". Journal of Zhejiang University. Science. B. 19 (10): 739–749. doi:10.1631/jzus.B1700594. PMC 6194357. PMID 30269442.
- Farooqi AA, Rehman ZU, Muntane J (November 2014). "Antisense therapeutics in oncology: current status". OncoTargets and Therapy. 7: 2035–42. doi:10.2147/ott.s49652. PMC 4224095. PMID 25395862.
- "Antisense Oligonucleotide-based Therapeutics", Gene Therapy, CRC Press, pp. 365–392, May 2000, ISBN 978-0-429-13193-6
- "Volume 49, 2009". Annual Review of Pharmacology and Toxicology. 49. 2009-01-10. doi:10.1146/pharmtox.2009.49.issue-1.
- Hanna J, Hossain GS, Kocerha J (2019). "The Potential for microRNA Therapeutics and Clinical Research". Frontiers in Genetics. 10: 478. doi:10.3389/fgene.2019.00478. PMC 6532434. PMID 31156715.
- Devi GR (September 2006). "siRNA-based approaches in cancer therapy". Cancer Gene Therapy. 13 (9): 819–29. doi:10.1038/sj.cgt.7700931. PMID 16424918. S2CID 23475841.
- Napoli C, Lemieux C, Jorgensen R (April 1990). "Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans". The Plant Cell. 2 (4): 279–289. doi:10.1105/tpc.2.4.279. PMC 159885. PMID 12354959.
- Hannon GJ (July 2002). "RNA interference". Nature. 418 (6894): 244–51. Bibcode:2002Natur.418..244H. doi:10.1038/418244a. PMID 12110901. S2CID 4426281.
- Tomari Y, Zamore PD (March 2005). "Perspective: machines for RNAi". Genes & Development. 19 (5): 517–29. doi:10.1101/gad.1284105. PMID 15741316.
- Banan M, Puri N (October 2004). "The ins and outs of RNAi in mammalian cells". Current Pharmaceutical Biotechnology. 5 (5): 441–50. doi:10.2174/1389201043376643. PMID 15544492.
- Song E, Lee SK, Dykxhoorn DM, Novina C, Zhang D, Crawford K, et al. (July 2003). "Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages". Journal of Virology. 77 (13): 7174–81. doi:10.1128/JVI.77.13.7174-7181.2003. PMC 164789. PMID 12805416.
- Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, et al. (March 2003). "RNA interference targeting Fas protects mice from fulminant hepatitis". Nature Medicine. 9 (3): 347–51. doi:10.1038/nm828. PMID 12579197. S2CID 4182632.
- Lai EC (April 2002). "Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation". Nature Genetics. 30 (4): 363–4. doi:10.1038/ng865. PMID 11896390. S2CID 33175420.
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, et al. (June 2003). "Expression profiling reveals off-target gene regulation by RNAi". Nature Biotechnology. 21 (6): 635–7. doi:10.1038/nbt831. PMID 12754523. S2CID 24778534.
- Bader AG, Brown D, Winkler M (September 2010). "The promise of microRNA replacement therapy". Cancer Research. 70 (18): 7027–30. doi:10.1158/0008-5472.CAN-10-2010. PMC 2940943. PMID 20807816.
- Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG (July 2010). "Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34". Cancer Research. 70 (14): 5923–30. doi:10.1158/0008-5472.CAN-10-0655. PMC 2913706. PMID 20570894.
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. (June 2009). "Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model". Cell. 137 (6): 1005–17. doi:10.1016/j.cell.2009.04.021. PMC 2722880. PMID 19524505.
- Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, et al. (August 2014). "A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore". Nature Chemical Biology. 10 (8): 686–91. doi:10.1038/nchembio.1561. PMC 4104137. PMID 24952597.
- Zhou J, Rossi J (March 2017). "Aptamers as targeted therapeutics: current potential and challenges". Nature Reviews. Drug Discovery. 16 (3): 181–202. doi:10.1038/nrd.2016.199. PMC 5700751. PMID 27807347.
- Guo P (December 2010). "The emerging field of RNA nanotechnology". Nature Nanotechnology. 5 (12): 833–42. Bibcode:2010NatNa...5..833G. doi:10.1038/nnano.2010.231. PMC 3149862. PMID 21102465.
- Gelinas AD, Davies DR, Janjic N (February 2016). "Embracing proteins: structural themes in aptamer-protein complexes". Current Opinion in Structural Biology. 36: 122–32. doi:10.1016/j.sbi.2016.01.009. PMID 26919170.
- Geiger A, Burgstaller P, von der Eltz H, Roeder A, Famulok M (March 1996). "RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity". Nucleic Acids Research. 24 (6): 1029–36. doi:10.1093/nar/24.6.1029. PMC 145747. PMID 8604334.
- Chen L, Rashid F, Shah A, Awan HM, Wu M, Liu A, et al. (August 2015). "The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation". Proceedings of the National Academy of Sciences of the United States of America. 112 (32): 10002–7. Bibcode:2015PNAS..11210002C. doi:10.1073/pnas.1502159112. PMC 4538674. PMID 26216949.
- Keefe AD, Pai S, Ellington A (July 2010). "Aptamers as therapeutics". Nature Reviews. Drug Discovery. 9 (7): 537–50. doi:10.1038/nrd3141. PMC 7097324. PMID 20592747.
- Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, et al. (February 2014). "Stable RNA nanoparticles as potential new generation drugs for cancer therapy". Advanced Drug Delivery Reviews. 66: 74–89. doi:10.1016/j.addr.2013.11.006. PMC 3955949. PMID 24270010.
- Tuerk C, Gold L (August 1990). "Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase". Science. 249 (4968): 505–10. Bibcode:1990Sci...249..505T. doi:10.1126/science.2200121. PMID 2200121.
- Ellington AD, Szostak JW (August 1990). "In vitro selection of RNA molecules that bind specific ligands". Nature. 346 (6287): 818–22. Bibcode:1990Natur.346..818E. doi:10.1038/346818a0. PMID 1697402. S2CID 4273647.
- Guo KT, Ziemer G, Paul A, Wendel HP (April 2008). "CELL-SELEX: Novel perspectives of aptamer-based therapeutics". International Journal of Molecular Sciences. 9 (4): 668–78. doi:10.3390/ijms9040668. PMC 2635693. PMID 19325777.
- Ohuchi S (2012). "Cell-SELEX Technology". BioResearch Open Access. 1 (6): 265–272. doi:10.1089/biores.2012.0253. PMC 3559206. PMID 23515081.
- Cerchia L, Giangrande PH, McNamara JO, de Franciscis V (2009). "Cell-specific aptamers for targeted therapies". In Mayer G (ed.). Nucleic Acid and Peptide Aptamers. Methods in Molecular Biology. Vol. 535. Totowa, NJ: Humana Press. pp. 59–78. doi:10.1007/978-1-59745-557-2_5. ISBN 978-1-59745-557-2. PMC 4443708. PMID 19377980.
- Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, et al. (July 2007). "Selection of aptamers for molecular recognition and characterization of cancer cells". Analytical Chemistry. 79 (13): 4900–7. doi:10.1021/ac070189y. PMID 17530817.
- Xiao Z, Shangguan D, Cao Z, Fang X, Tan W (2008-02-18). "Cell-specific internalization study of an aptamer from whole cell selection". Chemistry. 14 (6): 1769–75. doi:10.1002/chem.200701330. PMID 18092308.
- Phillips JA, Lopez-Colon D, Zhu Z, Xu Y, Tan W (July 2008). "Applications of aptamers in cancer cell biology". Analytica Chimica Acta. 621 (2): 101–8. doi:10.1016/j.aca.2008.05.031. PMID 18573375.
- Kohn DB, Bauer G, Rice CR, Rothschild JC, Carbonaro DA, Valdez P, et al. (July 1999). "A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children". Blood. 94 (1): 368–71. doi:10.1182/blood.V94.1.368.413a47_368_371. PMID 10381536.
- Khedri M, Rafatpanah H, Abnous K, Ramezani P, Ramezani M (December 2015). "Cancer immunotherapy via nucleic acid aptamers". International Immunopharmacology. 29 (2): 926–936. doi:10.1016/j.intimp.2015.10.013. PMID 26603636.
- Shi H, He X, Wang K, Wu X, Ye X, Guo Q, et al. (March 2011). "Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration". Proceedings of the National Academy of Sciences of the United States of America. 108 (10): 3900–5. Bibcode:2011PNAS..108.3900S. doi:10.1073/pnas.1016197108. PMC 3054025. PMID 21368158.
- Zhang J, Smaga LP, Satyavolu NS, Chan J, Lu Y (December 2017). "DNA Aptamer-Based Activatable Probes for Photoacoustic Imaging in Living Mice". Journal of the American Chemical Society. 139 (48): 17225–17228. doi:10.1021/jacs.7b07913. PMC 5724028. PMID 29028325.
- Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, Clary BM (January 2010). "In vivo selection of tumor-targeting RNA motifs". Nature Chemical Biology. 6 (1): 22–4. doi:10.1038/nchembio.277. PMC 2795795. PMID 19946274.
- Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, et al. (August 2006). "Aptamers evolved from live cells as effective molecular probes for cancer study". Proceedings of the National Academy of Sciences of the United States of America. 103 (32): 11838–43. doi:10.1073/pnas.0602615103. PMC 1567664. PMID 16873550.
- Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, et al. (June 2008). "Molecular recognition of small-cell lung cancer cells using aptamers". ChemMedChem. 3 (6): 991–1001. doi:10.1002/cmdc.200800030. PMC 3544301. PMID 18338423.
- Gragoudas ES, Adamis AP, Cunningham ET, Feinsod M, Guyer DR (December 2004). "Pegaptanib for neovascular age-related macular degeneration". The New England Journal of Medicine. 351 (27): 2805–16. doi:10.1056/NEJMoa042760. PMID 15625332. S2CID 10050130.
- Liang C, Guo B, Wu H, Shao N, Li D, Liu J, et al. (March 2015). "Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy". Nature Medicine. 21 (3): 288–94. doi:10.1038/nm.3791. PMC 5508976. PMID 25665179.
- Hoellenriegel J, Zboralski D, Maasch C, Rosin NY, Wierda WG, Keating MJ, et al. (February 2014). "The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization". Blood. 123 (7): 1032–9. doi:10.1182/blood-2013-03-493924. PMC 4123413. PMID 24277076.
- Mousa SA, Mousa SS (June 2010). "Current status of vascular endothelial growth factor inhibition in age-related macular degeneration". BioDrugs. 24 (3): 183–94. doi:10.2165/11318550-000000000-00000. PMID 20210371. S2CID 19177321.
- Ferrara N, Adamis AP (June 2016). "Ten years of anti-vascular endothelial growth factor therapy". Nature Reviews. Drug Discovery. 15 (6): 385–403. doi:10.1038/nrd.2015.17. PMID 26775688. S2CID 29441279.
- Healy JM, Lewis SD, Kurz M, Boomer RM, Thompson KM, Wilson C, McCauley TG (December 2004). "Pharmacokinetics and biodistribution of novel aptamer compositions". Pharmaceutical Research. 21 (12): 2234–46. doi:10.1007/s11095-004-7676-4. PMID 15648255. S2CID 25786376.
- Abeydeera ND, Egli M, Cox N, Mercier K, Conde JN, Pallan PS, et al. (September 2016). "Evoking picomolar binding in RNA by a single phosphorodithioate linkage". Nucleic Acids Research. 44 (17): 8052–64. doi:10.1093/nar/gkw725. PMC 5041495. PMID 27566147.
- Henry SP, Giclas PC, Leeds J, Pangburn M, Auletta C, Levin AA, Kornbrust DJ (May 1997). "Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: potential mechanism of action". The Journal of Pharmacology and Experimental Therapeutics. 281 (2): 810–6. PMID 9152389.
- Farman CA, Kornbrust DJ (2016-11-17). "Oligodeoxynucleotide studies in primates: antisense and immune stimulatory indications". Toxicologic Pathology. 31 Suppl: 119–22. doi:10.1080/01926230390174995. PMID 12597439. S2CID 22520922.
- Avci-Adali M, Steinle H, Michel T, Schlensak C, Wendel HP (2013-07-23). "Potential capacity of aptamers to trigger immune activation in human blood". PLOS ONE. 8 (7): e68810. Bibcode:2013PLoSO...868810A. doi:10.1371/journal.pone.0068810. PMC 3720859. PMID 23935890.
- Mena A, Nichani AK, Popowych Y, Godson DL, Dent D, Townsend HG, et al. (October 2003). "Innate immune responses induced by CpG oligodeoxyribonucleotide stimulation of ovine blood mononuclear cells". Immunology. 110 (2): 250–7. doi:10.1046/j.1365-2567.2003.01722.x. PMC 1783041. PMID 14511239.
- Musumeci D, Montesarchio D (November 2012). "Polyvalent nucleic acid aptamers and modulation of their activity: a focus on the thrombin binding aptamer". Pharmacology & Therapeutics. 136 (2): 202–15. doi:10.1016/j.pharmthera.2012.07.011. PMID 22850531.
- Soule EE, Bompiani KM, Woodruff RS, Sullenger BA (February 2016). "Targeting Two Coagulation Cascade Proteases with a Bivalent Aptamer Yields a Potent and Antidote-Controllable Anticoagulant". Nucleic Acid Therapeutics. 26 (1): 1–9. doi:10.1089/nat.2015.0565. PMC 4753633. PMID 26584417.
- Ganson NJ, Povsic TJ, Sullenger BA, Alexander JH, Zelenkofske SL, Sailstad JM, et al. (May 2016). "Pre-existing anti-polyethylene glycol antibody linked to first-exposure allergic reactions to pegnivacogin, a PEGylated RNA aptamer". The Journal of Allergy and Clinical Immunology. 137 (5): 1610–1613.e7. doi:10.1016/j.jaci.2015.10.034. PMC 5819876. PMID 26688515.
- Lincoff AM, Mehran R, Povsic TJ, Zelenkofske SL, Huang Z, Armstrong PW, et al. (January 2016). "Effect of the REG1 anticoagulation system versus bivalirudin on outcomes after percutaneous coronary intervention (REGULATE-PCI): a randomised clinical trial". Lancet. 387 (10016): 349–356. doi:10.1016/s0140-6736(15)00515-2. PMID 26547100. S2CID 10905525.
- Sharma TK, Bruno JG, Dhiman A (2017-03-01). "ABCs of DNA aptamer and related assay development". Biotechnology Advances. 35 (2): 275–301. doi:10.1016/j.biotechadv.2017.01.003. PMID 28108354.
- Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F (July 1991). "Kinetic characterization of ribonuclease-resistant 2'-modified hammerhead ribozymes". Science. 253 (5017): 314–7. Bibcode:1991Sci...253..314P. doi:10.1126/science.1857967. PMID 1857967.
- Vater A, Klussmann S (March 2003). "Toward third-generation aptamers: Spiegelmers and their therapeutic prospects". Current Opinion in Drug Discovery & Development. 6 (2): 253–61. PMID 12669461.
- Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G, et al. (November 2004). "Antidote-mediated control of an anticoagulant aptamer in vivo". Nature Biotechnology. 22 (11): 1423–8. doi:10.1038/nbt1023. PMID 15502817. S2CID 5061714.
- Aaldering LJ, Tayeb H, Krishnan S, Fletcher S, Wilton SD, Veedu RN (2015-04-03). "Smart functional nucleic acid chimeras: enabling tissue specific RNA targeting therapy". RNA Biology. 12 (4): 412–25. doi:10.1080/15476286.2015.1017234. PMC 4615226. PMID 25849197.
External links
- RNA therapeutics on the rise, Nature (April 2020)
.jpg.webp)