TCF21 (gene)
Transcription factor 21 (TCF21), also known as pod-1, capsuling, or epicardin, is a protein that in humans is encoded by the TCF21 gene on chromosome 6.[5][6][7] It is ubiquitously expressed in many tissues and cell types and highly significantly expressed in lung and placenta.[8] TCF21 is crucial for the development of a number of cell types during embryogenesis of the heart,[9] lung,[10] kidney,[10] and spleen.[11] TCF21 is also deregulated in several types of cancers, and thus known to function as a tumor suppressor.[12] The TCF21 gene also contains one of 27 SNPs associated with increased risk of coronary artery disease.[13]
Discovery
TCF21 was discovered in 1998 when search for novel cell-type-specific bHLH proteins expressed in human and mouse kidneys by performing a search of the expressed sequence tag (EST) databases.[6] Because the transcript they found was highly expressed in visceral glomerular epithelial cells (podocytes), TCF21 was initialled named Pod-1. Comparison of Pod-1 with previously characterized bHLH proteins identified Pod-1 as a novel member of a subfamily of bHLH proteins with important roles in mesodermal development.[6] The chromosomal location of Pod-1 in the mouse was then determined using an interspecific backcross panel along with genomic southern blot analysis to identify restriction fragment length polymorphisms (RFLPs) between inbred mouse strains. Analysis showed Pod-1 to map to a region of mouse chromosome 10 that is syntenic with human chromosome 6q23-q24.[6]
The tissue distribution of Pod-1 was determined by hybridization of a human multiple tissue northern blot with a Pod-1 cDNA. A probe lacking the bHLH domain was used to minimize cross-reactivity along with high stringency hybridization and washing. Results showed that in humans and mice, Pod-1 was most highly expressed in the kidney, lung and heart, with selective expression at sites of epithelial-mesenchymal interaction in the kidney, lung, intestine and pancreas of developing mouse embryos.[6] RNA in situ hybridization using 33P-labeled riboprobes was used to identify the cell types that expressed Pod-1 in the developing kidney and other tissues. This revealed Pod-1 expression in mesenchymal cells and podocytes, with expression coinciding with the onset of podocyte differentiation.[6] It was found that expression of Pod-1 in embryonic kidney explants was inhibited through antisense oligonucleotides. This inhibition resulted in decreased mesenchymal cell condensation around the ureteric bud and a significant decrease in ureteric branching. Pod-1 was the first tissue-restricted bHLH protein to be identified in the developing kidney and tied to regulation of morphogenetic events.[6]
In an effort to identify novel bHLH factors related to dHAND and eHAND (a novel subclass of cell type-restricted bHLH factors shown to play important roles in cardiac morphogenesis), they screened expressed sequence tag (EST) databases for sequences with homology to the bHLH regions of these factors.[14] The novel bHLH protein they identified in their search was also Pod-1, but they used the name capsulin.
Whole-mount in situ hybridization of Capsulin transcripts were used to define sites of expression, which showed to be specific to mesodermal precursor cells that surround the epithelium of the developing gastrointestinal, genitourinary and respiratory systems during mouse embryogenesis.[15] Expression patterns of capsulin mRNA in adult mouse tissues by Northern blot detected highest levels in the lungs, with lower levels in kidneys, heart and spleen. Capsulin transcripts were also found to mark the spiral septum of the heart and progenitor cells that give rise to the pericardium and coronary arteries.[15]
Capsulin was translated in a rabbit reticulocyte lysate in the presence and absence of the widely expressed bHLH protein E12 and performed gel mobility shift assays with several E-box sequences as probes to test the protein's DNA binding activity.[15] Capsulin alone failed to bind any of the sequences tested. However, in the presence of E12 plus capsulin, a DNA complex was generated with the probe that migrated faster than the E12 homodimeric complex alone, representing the binding of capsulin/E12 heterodimers.[15] It was concluded that capsulin heterodimerizes with E12 and binds the specific E-box consensus sequence (CANNTG), though not activating transcription through this sequence on its own. Its restricted expression pattern and DNA binding activity identified Capsulin as a regulator of gene expression in specific subtypes of visceral mesodermal cells involved in organogenesis and in precursor cells that contribute to the pericardium, coronary arteries and regions of the heart.[15]
Structure
Gene
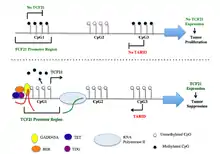
The TCF21 gene resides on chromosome 6 at the band 6q23.2 and includes 3 exons.[7] These three exons are associated with CpG islands CGI1, CGI2, and CGI3. DNA methylation analysis revealed hypermethylation at CGI1 and CGI3, but not CGI2 in samples from various cancer tissues.[16] Luciferase reporter assays with constructs covering CGI3 sequences in sense and antisense orientation demonstrated that CGI3 harbors a promoter that directs the synthesis of a previously unknown long non-coding RNAs (lncRNAs) in antisense orientation to TCF21. This lncRNA has been named TARID (for TCF21 antisense RNA inducing demethylation).[17]
Function
TCF21 encodes a transcription factor of the basic helix-loop-helix (bHLH) family, which manages cell-fate specification, commitment and differentiation in various cell lineages during development.[9] The TCF21 product is mesoderm-specific and expressed in embryonic epicardium, mesenchyme-derived tissues of lung, gut, gonad, and both mesenchymal and glomerular epithelial cells in the kidney.[7]
TCF21 is essential for regulating properties of the mesenchyme that are critically important for several aspects of lung and kidney morphogenesis.[10] TCF21 is also essential for cardiac fibroblast cell fate, as demonstrated by the failed development of cardiac fibroblasts in mice lacking TCF21.[9] In the absence of TCF21, these fibroblast progenitor cells do not undergo epithelial-to-mesenchymal transition (EMT). While TCF21-expressing epicardial cells are initially multipotent, they become committed to the fibroblast lineage over time. Those TCF21-expressing cells that do not commit to the fibroblast lineage lose this expression and remain undifferentiated epicardial cells or coronary vascular smooth muscle cells.[9] TCF21 is expressed in mesodermal cells in the proepicardial organ that give rise to coronary artery smooth muscle cells (SMC) and loss of TCF21 results in increased expression of smooth muscle markers by cells on the heart surface consistent with premature SMC differentiation. This suggests that early expression of TCF21 is important for expansion of the SMC compartment of the coronary circulation, with persistent TCF21 expression being required for cardiac fibroblast development.[20]
TCF21 activation is directed by an antisense long non-coding RNA, TARID (TCF21 antisense RNA inducing demethylation). TARID activates TCF21 expression by inducing promoter demethylation and affects expression levels of target genes by functioning as epigenetic regulators of chromatin structure through interactions with histone modifiers, chromatin remodeling complexes, transcriptional regulators, and/or DNA methylation machinery.[17]
Role in development
Since the identification of Tcf21 significance in various cell lineages, further research has expanded understanding of the essential roles of this gene.
TCF21 is essential for regulating properties of the mesenchyme that are critically important for several aspects of lung and kidney morphogenesis. Null TCF21 mutant mice are born but die shortly after due to severely hypoplastic lungs and kidneys that lack alveoli and mature glomeruli.[10] While TCF21 is exclusively expressed in the mesenchyme and podocytes, major defects are observed in adjacent epithelia of TCF21 mutant mice. In the kidney, TCF21 is required for conversion of condensing mesenchyme to epithelium of the nephron, branching morphogenesis and terminal differentiation of tubular epithelium. In the lung, TCF21 is required to correctly pattern the proximodistal axis of airway epithelium and for normal branching to occur.[10]
TCF21 null mice also fail to form a spleen, as TCF21 acts after splenic specification to control morphogenetic expansion of the splenic anlage and in its absence, splenic precursor cells undergo apoptosis.[11] Since this splenic phenotype resembles that of mice lacking the homeobox genes Hox11 and Bapx1, it is possible that TCF21, together with Hox11, and Bapx1 control a common essential early step in the developmental pathway for spleen organogenesis.[11]
TCF21 is essential for cardiac fibroblast cell fate, as demonstrated by the failed development of cardiac fibroblasts in mice lacking TCF21.[9] In the absence of TCF21, these fibroblast progenitor cells do not undergo epithelial-to-mesenchymal transition (EMT). While TCF21-expressing epicardial cells are initially multipotent, they become committed to the fibroblast lineage over time. Those TCF21-expressing cells that do not commit to the fibroblast lineage lose this expression and remain undifferentiated epicardial cells or coronary vascular smooth muscle cells.[9] TCF21 is expressed in mesodermal cells in the proepicardial organ that give rise to coronary artery smooth muscle cells (SMC) and loss of TCF21 results in increased expression of smooth muscle markers by cells on the heart surface consistent with premature SMC differentiation. This suggests that early expression of TCF21 is important for expansion of the SMC compartment of the coronary circulation, with persistent TCF21 expression being required for cardiac fibroblast development.[20]
Male TCF21 knockout mice, which die at birth due to respiratory failure, are reported to have feminized genitalia, implicating TCF21 in mouse gonadal development/differentiation.[21] TCF21 transcriptionally represses steroidogenic factor 1 (Sf1), a gene expression regulator that mediates sexual differentiation and is involved in coordinating cell fate decisions in gonadal progenitors.[21] Without TCF21, normal gonad development is disrupted as a result of ectopic expression of Sf1, which leads to abnormal committing of urogenital progenitor cells to steroidogenic cell fates. In the XY gonad, this disruption in organization contributes to changes in testicular structure and vasculature.[21]
Clinical significance
As a cancer suppressor
In humans, TCF21 has been identified as a candidate tumor suppressor gene and is frequently epigenetically silenced in various human cancers.
Restriction landmark genomic scanning (RLGS) along a region of recurrent loss of heterozygosity (LOH) at chromosome 6q23-q24 to profile DNA methylation was used to test the hypothesis that abnormal promoter methylation could help pinpoint the location of a candidate tumor suppressor in regions of LOH. 6q23-q24 was the chosen chromosomal region due to frequently described LOH in human head and neck squamous cell carcinomas (HNSCC) and non-small-cell lung cancers (NSCLC) as well as in other tumor types, but with no identified tumor suppressor.[12] Hypermethylation was found to occur frequently in the same RLGS loci in HNSCC and NSCLC. Sodium bisulfite sequencing further identified tumor-specific methylation of TCF21 when compared to normal controls. RNA samples were isolated from tumor tissues and analyzed to correlate the amount of TCF21 mRNA and DNA methylation in the samples. Overall, tumor samples with higher levels of CpG island hypermethylation had decreased TCF21 expression than normal controls.[12] Exogenous expression of TCF21 in cells with silenced endogenous TCF21 loci resulted in a reductions in tumor properties both in vitro and in vivo. Based on these results, it was concluded that TCF21 is a tumor suppressor gene, often silenced by hypermethylation in cancer.[12]
TCF21 has also been linked to metastatic melanoma progression through the inhibition of the KISS1 metastasis-suppressor gene.[22] DNA methylation analysis in melanoma patient biopsies has demonstrated downregulation of TCF21 due to promoter hypermethylation, which also correlates with decreased survival in patients suffering from metastatic skin melanoma.[22] TCF21, together with E12 and TCF12, bind the KISS1 promoter, sustaining its activity. Without TCF21 to interact with the KISS1 promoter, KISS1 expression is lost. Melanoma cells overexpressing TCF21 have also been found to display reduced motility compared with vector-only control cells.[22]
Tobacco-induced lung cancer research has found TCF21 to be among the genes identified as highly methylated at both high and low concentrations of cigarette smoke condensate (CSC). In the presence genistein, one of the soy-derived bioactive isoflavones, methylation of TCF21 is significantly reduced.[23] Genistein has been known to affect tumorigenesis through epigenetic regulation, such as chromatin configuration and DNA methylation, activating other tumor suppressor genes that affect cancer cell survival.[24] These findings support the hypothesis that increasing hypermethylated tumor suppressor genes such as TCF21 is a potential chemopreventative pathway in tobacco-induced lung cancer.[23]
TCF21 may have also therapeutic potential for breast cancer treatment, as downregulation of TCF21 has been implicated in breast cancer tumorigenesis and proliferation.[15] TCF21 mRNA expression is very low in breast cancer cells compared with normal breast epithelial cells. This low expression is also associated with large tumor size and lymph node metastasis. Breast cancer tissues exhibit significantly downregulated expression of TCF21 mRNA and TCF21 mRNA overexpression has been found to inhibit cancerous cell proliferation.[15]
Clinical marker
TCF21 methylation has been considered as a potential clinical marker in the diagnosis of renal cell carcinoma.[25] Accordingly, TCF21 methylation levels in urine samples may be a useful means of diagnosing renal cell carcinoma. Also TCF21 rs12190287 polymorphism can regulate TCF21 expression and may serve as a potential marker for genetic susceptibility to breast cancer.[26]
Additionally, a multi-locus genetic risk score study based on a combination of 27 loci, including the TCF21 gene, identified individuals at increased risk for both incident and recurrent coronary artery disease events, as well as an enhanced clinical benefit from statin therapy. The study was based on a community cohort study (the Malmo Diet and Cancer study) and four additional randomized controlled trials of primary prevention cohorts (JUPITER and ASCOT) and secondary prevention cohorts (CARE and PROVE IT-TIMI 22).[27]
References
- GRCh38: Ensembl release 89: ENSG00000118526 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000045680 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Lotfi CF, Passaia BS, Kremer JL (2021). "Role of the bHLH transcription factor TCF21 in development and tumorigenesis". Brazilian Journal of Medical and Biological Research. 54 (5): e10637. doi:10.1590/1414-431X202010637. PMC 7959166. PMID 33729392.
- Quaggin SE, Vanden Heuvel GB, Igarashi P (February 1998). "Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney". Mechanisms of Development. 71 (1–2): 37–48. doi:10.1016/S0925-4773(97)00201-3. PMID 9507058. S2CID 17400923.
- "Entrez Gene: TCF21 transcription factor 21".
- "BioGPS - your Gene Portal System". biogps.org. Retrieved 11 October 2016.
- Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, Olson EN, Tallquist MD (June 2012). "The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors". Development. 139 (12): 2139–49. doi:10.1242/dev.079970. PMC 3357908. PMID 22573622.
- Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J (December 1999). "The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis". Development. 126 (24): 5771–83. doi:10.1242/dev.126.24.5771. PMID 10572052.
- Lu J, Chang P, Richardson JA, Gan L, Weiler H, Olson EN (August 2000). "The basic helix-loop-helix transcription factor capsulin controls spleen organogenesis". Proceedings of the National Academy of Sciences of the United States of America. 97 (17): 9525–30. doi:10.1073/pnas.97.17.9525. PMC 16898. PMID 10944221.
- Smith LT, Lin M, Brena RM, Lang JC, Schuller DE, Otterson GA, Morrison CD, Smiraglia DJ, Plass C (January 2006). "Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer". Proceedings of the National Academy of Sciences of the United States of America. 103 (4): 982–7. Bibcode:2006PNAS..103..982S. doi:10.1073/pnas.0510171102. PMC 1348006. PMID 16415157.
- Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield MJ, Devlin JJ, et al. (June 2015). "Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials". Lancet. 385 (9984): 2264–71. doi:10.1016/S0140-6736(14)61730-X. PMC 4608367. PMID 25748612.
- Lu J, Richardson JA, Olson EN (April 1998). "Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs". Mechanisms of Development. 73 (1): 23–32. doi:10.1016/s0925-4773(98)00030-6. PMID 9545521. S2CID 14075581.
- Wang J, Gao X, Wang M, Zhang J (November 2015). "Clinicopathological significance and biological role of TCF21 mRNA in breast cancer". Tumour Biology. 36 (11): 8679–83. doi:10.1007/s13277-015-3476-1. PMID 26044559. S2CID 24587856.
- Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, Dienemann H, Dyckhoff G, Herold-Mende C, Grummt I, Niehrs C, Plass C (August 2014). "Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A". Molecular Cell. 55 (4): 604–14. doi:10.1016/j.molcel.2014.06.031. PMID 25087872.
- Rinn JL, Chang HY (2012). "Genome regulation by long noncoding RNAs". Annual Review of Biochemistry. 81: 145–66. doi:10.1146/annurev-biochem-051410-092902. PMC 3858397. PMID 22663078.
- "TCF21 - Transcription factor 21 - Homo sapiens (Human) - TCF21 gene & protein". www.uniprot.org. Retrieved 11 October 2016.
- Hidai H, Bardales R, Goodwin R, Quertermous T, Quertermous EE (April 1998). "Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries". Mechanisms of Development. 73 (1): 33–43. doi:10.1016/s0925-4773(98)00031-8. PMID 9545526. S2CID 18884227.
- Sazonova O, Zhao Y, Nürnberg S, Miller C, Pjanic M, Castano VG, Kim JB, Salfati EL, Kundaje AB, Bejerano G, Assimes T, Yang X, Quertermous T (May 2015). "Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci". PLOS Genetics. 11 (5): e1005202. doi:10.1371/journal.pgen.1005202. PMC 4447360. PMID 26020271.
- Cui S, Ross A, Stallings N, Parker KL, Capel B, Quaggin SE (August 2004). "Disrupted gonadogenesis and male-to-female sex reversal in Pod1 knockout mice". Development. 131 (16): 4095–105. doi:10.1242/dev.01266. PMID 15289436.
- Arab K, Smith LT, Gast A, Weichenhan D, Huang JP, Claus R, Hielscher T, Espinosa AV, Ringel MD, Morrison CD, Schadendorf D, Kumar R, Plass C (October 2011). "Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma". Carcinogenesis. 32 (10): 1467–73. doi:10.1093/carcin/bgr138. PMC 3179423. PMID 21771727.
- Lyn-Cook L, Word B, George N, Lyn-Cook B, Hammons G (2014). "Effect of cigarette smoke condensate on gene promoter methylation in human lung cells". Tobacco Induced Diseases. 12 (1): 15. doi:10.1186/1617-9625-12-15. PMC 4160916. PMID 25214829.
- Zhang Y, Chen H (July 2011). "Genistein, an epigenome modifier during cancer prevention". Epigenetics. 6 (7): 888–91. doi:10.4161/epi.6.7.16315. PMID 21610327.
- Xin J, Xu R, Lin S, Xin M, Cai W, Zhou J, Fu C, Zhen G, Lai J, Li Y, Zhang P (August 2016). "Clinical potential of TCF21 methylation in the diagnosis of renal cell carcinoma". Oncology Letters. 12 (2): 1265–1270. doi:10.3892/ol.2016.4748. PMC 4950740. PMID 27446425.
- Gao X, Yang J, Wang M, Zhang J (June 2016). "TCF21 genetic polymorphisms and breast cancer risk in Chinese women". Oncotarget. 7 (34): 55757–55764. doi:10.18632/oncotarget.9825. PMC 5342451. PMID 27270650.
- Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield MJ, Devlin JJ, Nordio F, Hyde CL, Cannon CP, Sacks FM, Poulter NR, Sever PS, Ridker PM, Braunwald E, Melander O, Kathiresan S, Sabatine MS (June 2015). "Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials". Lancet. 385 (9984): 2264–71. doi:10.1016/S0140-6736(14)61730-X. PMC 4608367. PMID 25748612.
Further reading
- Sazonova O, Zhao Y, Nürnberg S, Miller C, Pjanic M, Castano VG, Kim JB, Salfati EL, Kundaje AB, Bejerano G, Assimes T, Yang X, Quertermous T (May 2015). "Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci". PLOS Genetics. 11 (5): e1005202. doi:10.1371/journal.pgen.1005202. PMC 4447360. PMID 26020271.
- Littlewood TD, Evan GI (1995). "Transcription factors 2: helix-loop-helix". Protein Profile. 2 (6): 621–702. PMID 7553065.
- Bonaldo MF, Lennon G, Soares MB (September 1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Research. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Hidai H, Bardales R, Goodwin R, Quertermous T, Quertermous EE (April 1998). "Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries". Mechanisms of Development. 73 (1): 33–43. doi:10.1016/S0925-4773(98)00031-8. PMID 9545526. S2CID 18884227.
- Robb L, Mifsud L, Hartley L, Biben C, Copeland NG, Gilbert DJ, Jenkins NA, Harvey RP (September 1998). "epicardin: A novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads". Developmental Dynamics. 213 (1): 105–13. doi:10.1002/(SICI)1097-0177(199809)213:1<105::AID-AJA10>3.0.CO;2-1. PMID 9733105. S2CID 25946118.
- Suzuki H, Fukunishi Y, Kagawa I, Saito R, Oda H, Endo T, Kondo S, Bono H, Okazaki Y, Hayashizaki Y (October 2001). "Protein-protein interaction panel using mouse full-length cDNAs". Genome Research. 11 (10): 1758–65. doi:10.1101/gr.180101. PMC 311163. PMID 11591653.
- Funato N, Ohyama K, Kuroda T, Nakamura M (February 2003). "Basic helix-loop-helix transcription factor epicardin/capsulin/Pod-1 suppresses differentiation by negative regulation of transcription". The Journal of Biological Chemistry. 278 (9): 7486–93. doi:10.1074/jbc.M212248200. PMID 12493738.
- Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, Meil A, Wojcik J, Legrain P, Gauthier JM (July 2004). "Functional proteomics mapping of a human signaling pathway". Genome Research. 14 (7): 1324–32. doi:10.1101/gr.2334104. PMC 442148. PMID 15231748.