Interbreeding between archaic and modern humans
There is evidence for interbreeding between archaic and modern humans during the Middle Paleolithic and early Upper Paleolithic. The interbreeding happened in several independent events that included Neanderthals and Denisovans, as well as several unidentified hominins.[2]
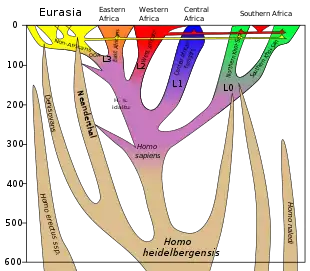
In Eurasia, interbreeding between Neanderthals and Denisovans with modern humans took place several times. The introgression events into modern humans are estimated to have happened about 47,000–65,000 years ago with Neanderthals and about 44,000–54,000 years ago with Denisovans.
Neanderthal-derived DNA has been found in the genomes of most or possibly all contemporary populations, varying noticeably by region. It accounts for 1–4% of modern genomes for people outside Sub-Saharan Africa, although estimates vary, and either none or possibly up to 0.3% — according to recent research[3] — for those in Africa. It is highest in East Asians, intermediate in Europeans, and lower in Southeast Asians.[4] According to some research, it is also lower in Melanesians compared to both East Asians and Europeans.[4] However, other research finds higher Neanderthal admixture in Melanesians, as well as in Native Americans, than in Europeans (though not higher than in East Asians).[5]
Denisovan-derived ancestry is largely absent from modern populations in Africa and Western Eurasia. The highest rates, by far, of Denisovan admixture have been found in Oceanian and some Southeast Asian populations. An estimated 4–6% of the genome of modern Melanesians is derived from Denisovans, but the highest amounts detected thus far are found in the Negrito populations of the Philippines. While some Southeast Asian Negrito populations carry Denisovan admixture, others have none, such as the Andamanese. In addition, low traces of Denisovan-derived ancestry have been found in mainland Asia, with an elevated Denisovan ancestry in South Asian populations compared to other mainland populations.
In Africa, archaic alleles consistent with several independent admixture events in the subcontinent have been found. It is currently unknown who these archaic African hominins were.[4]
Introgression of DNA from other lineages has enabled humanity "to migrate to, and succeed in, numerous new environments". Therefore, a strong case can be made that hybridization was an essential driving force in the emergence of modern humans.[6]
Neanderthals
Genetics
Proportion of admixture

On 7 May 2010, following the genome sequencing of three Vindija Neanderthals, a draft sequence of the Neanderthal genome was published and revealed that Neanderthals shared more alleles with Eurasian populations (e.g. French, Han Chinese, and Papua New Guinean) than with sub-Saharan African populations (e.g. Yoruba and San).[8] According to Green et al. (2010), the authors, the observed excess of genetic similarity is best explained by recent gene flow from Neanderthals to modern humans after the migration out of Africa.[8] They estimated the proportion of Neanderthal-derived ancestry to be 1–4% of the Eurasian genome.[8] Prüfer et al. (2013) estimated the proportion to be 1.5–2.1% for non-Africans,[9] Lohse and Frantz (2014) infer a higher rate of 3.4–7.3% in Eurasia.[10] In 2017, Prüfer et al. revised their estimate to 1.8–2.6% for non-Africans outside Oceania.[11]
According to a later study by Chen et al. (2020), Africans (specifically, the 1000 Genomes African populations) also have Neanderthal admixture,[12] with this Neanderthal admixture in African individuals accounting for 17 megabases,[12] which is 0.3% of their genome.[3] According to the authors, Africans gained their Neanderthal admixture predominantly from a back-migration by peoples (modern humans carrying Neanderthal admixture) that had diverged from ancestral Europeans (postdating the split between East Asians and Europeans).[12] This back-migration is proposed to have happened about 20,000 years ago.[3] However, some scientists, such as geneticist David Reich, have doubts about how extensive the flow of DNA back to Africa would have been, finding the signal of Neanderthal admixture "really weak".[13]
Introgressed genome
About 20% of the Neanderthal genome has been found introgressed or assimilated in the modern human population (by analyzing East Asians and Europeans),[14] but the figure has also been estimated at about a third.[15]
Subpopulation admixture rate
A higher Neanderthal admixture was found in East Asians than in Europeans,[14][16][17][18][19] which is estimated to be about 20% more introgression into East Asians.[14][16][19] This could possibly be explained by the occurrence of further admixture events in the early ancestors of East Asians after the separation of Europeans and East Asians,[4][14][16][17][19] dilution of Neanderthal ancestry in Europeans by populations with low Neanderthal ancestry from later migrations,[4][16][19] or natural selection that may have been relatively lower in East Asians than in Europeans.[4][18][19] Studies simulating admixture models indicate that a reduced efficacy of purifying selection against Neanderthal alleles in East Asians could not account for the greater proportion of Neanderthal ancestry of East Asians, thus favoring more-complex models involving additional pulses of Neanderthal introgression into East Asians.[20][21] Such models show a pulse to ancestral Eurasians, followed by separation and an additional pulse to ancestral East Asians.[4] It is observed that there is a small but significant variation of Neanderthal admixture rates within European populations, but no significant variation within East Asian populations.[14] Prüfer et al. (2017) remarked that East Asians carry more Neanderthal DNA (2.3–2.6%) than Western Eurasians (1.8–2.4%).[11]
It was later determined by Chen et al. (2020) that East Asians have 8% more Neanderthal ancestry, revised from the previous reports of 20% more Neanderthal ancestry, compared to Europeans.[12] This stems from the fact that Neanderthal ancestry shared with Africans had been masked, because Africans were thought to have no Neanderthal admixture and were therefore used as reference samples.[12] Thus, any overlap in Neanderthal admixture with Africans resulted in an underestimation of Neanderthal admixture in non-Africans and especially in Europeans.[12] The authors give a single pulse of Neanderthal admixture after the out-of-Africa dispersal as the most parsimonious explanation for the enrichment in East Asians, but they add that variation in Neanderthal ancestry may also be attributed to dilution to account for the now-more-modest differences found.[12] As a proportion of the total amount of Neanderthal sequence for each population, 7.2% of the sequence in Europeans is shared exclusively with Africans, while 2% of the sequence in East Asians is shared exclusively with Africans.[12]
Genomic analysis suggests that there is a global division in Neanderthal introgression between sub-Saharan African populations and other modern human groups (including North Africans) rather than between African and non-African populations.[22] North African groups share a similar excess of derived alleles with Neanderthals as do non-African populations, whereas sub-Saharan African groups are the only modern human populations that generally did not experience Neanderthal admixture.[23] The Neanderthal genetic signal among North African populations was found to vary depending on the relative quantity of autochthonous North African, European, Near Eastern and sub-Saharan ancestry. Using f4 ancestry ratio statistical analysis, the Neanderthal inferred admixture was observed to be: highest among the North African populations with maximal autochthonous North African ancestry such as Tunisian Berbers, where it was at the same level or even higher than that of Eurasian populations (100–138%); high among North African populations carrying greater European or Near Eastern admixture, such as groups in North Morocco and Egypt (∼60–70%); and lowest among North African populations with greater Sub-Saharan admixture, such as in South Morocco (20%).[24] Quinto et al. (2012) therefore postulate that the presence of this Neanderthal genetic signal in Africa is not due to recent gene flow from Near Eastern or European populations since it is higher among populations bearing indigenous pre-Neolithic North African ancestry.[25] Low but significant rates of Neanderthal admixture has also been observed for the Maasai of East Africa.[26] After identifying African and non-African ancestry among the Maasai, it can be concluded that recent non-African modern human (post-Neanderthal) gene flow was the source of the contribution since around an estimated 30% of the Maasai genome can be traced to non-African introgression from about 100 generations ago.[17]
Distance to lineages

Presenting a high-quality genome sequence of a female Altai Neanderthal, it has been found that the Neanderthal component in non-African modern humans is more related to the Mezmaiskaya Neanderthal (North Caucasus) than to the Altai Neanderthal (Siberia) or the Vindija Neanderthals (Croatia).[9] By high-coverage sequencing the genome of a 50,000-year-old female Vindija Neanderthal fragment, it was later found that the Vindija and Mezmaiskaya Neanderthals did not seem to differ in the extent of their allele-sharing with modern humans.[11] In this case, it was also found that the Neanderthal component in non-African modern humans is more closely related to the Vindija and Mezmaiskaya Neanderthals than to the Altai Neanderthal.[11] These results suggest that a majority of the admixture into modern humans came from Neanderthal populations that had diverged (about 80–100kya) from the Vindija and Mezmaiskaya Neanderthal lineages before the latter two diverged from each other.[11]
Analysis of chromosome 21 of the Altai, El Sidrón (Spain), and Vindija Neanderthals indicates that of these three lineages, only the El Sidrón and Vindija Neanderthals display significant rates of gene flow (0.3–2.6%) into modern humans, suggesting that the El Sidrón and Vindija Neanderthals are more closely related than the Altai Neanderthal to the Neanderthals that interbred with modern humans about 47,000–65,000 years ago.[28] Conversely, significant rates of modern human gene flow into Neanderthals occurred—of the three examined lineages—for only the Altai Neanderthal (0.1–2.1%), suggesting that modern human gene flow into Neanderthals mainly took place after the separation of the Altai Neanderthals from the El Sidrón and Vindija Neanderthals that occurred roughly 110,000 years ago.[28] The findings show that the source of modern human gene flow into Neanderthals originated from a population of early modern humans from about 100,000 years ago, predating the out-of-Africa migration of the modern human ancestors of present-day non-Africans.[28]
Mitochondrial DNA and Y chromosome
No evidence of Neanderthal mitochondrial DNA has been found in modern humans.[29][30][31] This suggests that successful Neanderthal admixture happened in pairings with Neanderthal males and modern human females.[32][33] Possible hypotheses are that Neanderthal mitochondrial DNA had detrimental mutations that led to the extinction of carriers, that the hybrid offspring of Neanderthal mothers were raised in Neanderthal groups and became extinct with them, or that female Neanderthals and male Sapiens did not produce fertile offspring.[32] However, the hypothesized incompatibility between Neanderthals and modern humans is contested by findings that suggest that the Y chromosome of Neanderthals was replaced by an extinct lineage of the modern human Y chromosome, which introgressed into Neanderthals between 100,000 and 370,000 years ago.[34] Furthermore, the study concludes that the replacement of the Y chromosomes and mitochondrial DNA in Neanderthals after gene flow from modern humans is highly plausible given the increased genetic load in Neanderthals relative to modern humans.[34]
As shown in an interbreeding model produced by Neves and Serva (2012), the Neanderthal admixture in modern humans may have been caused by a very low rate of interbreeding between modern humans and Neanderthals, with the exchange of one pair of individuals between the two populations in about every 77 generations.[35] This low rate of interbreeding would account for the absence of Neanderthal mitochondrial DNA from the modern human gene pool as found in earlier studies, as the model estimates a probability of only 7% for a Neanderthal origin of both mitochondrial DNA and Y chromosome in modern humans.[35]
Reduced contribution
There are large genomic regions with strongly reduced Neanderthal contribution in modern humans due to negative selection,[14][18] partly caused by hybrid male infertility.[18] These regions were most-pronounced on the X chromosome, with fivefold lower Neanderthal ancestry compared to autosomes.[4][18] They also contained relatively high numbers of genes specific to testes.[18] This means that modern humans have relatively few Neanderthal genes that are located on the X chromosome or expressed in the testes, suggesting male infertility as a probable cause.[18] It may be partly affected by hemizygosity of X chromosome genes in males.[4]
Deserts of Neanderthal sequences may also be caused by genetic drift involving intense bottlenecks in the modern human population and background selection as a result of strong selection against deleterious Neanderthal alleles.[4] The overlap of many deserts of Neanderthal and Denisovan sequences suggests that repeated loss of archaic DNA occur at specific loci.[4]
It has also been shown that Neanderthal ancestry has been selected against in conserved biological pathways, such as RNA processing.[18]
Consistent with the hypothesis that purifying selection has reduced Neanderthal contribution in present-day modern human genomes, Upper Paleolithic Eurasian modern humans (such as the Tianyuan modern human) carry more Neanderthal DNA (about 4–5%) than present-day Eurasian modern humans (about 1–2%).[36]
Rates of selection against Neanderthal sequences varied for European and Asian populations.[4]
Changes in modern humans
In Eurasia, modern humans have adaptive sequences introgressed from archaic humans, which provided a source of advantageous genetic variants that are adapted to local environments and a reservoir for additional genetic variation.[4] Adaptive introgression from Neanderthals has targeted genes involved with keratin filaments, sugar metabolism, muscle contraction, body fat distribution, enamel thickness, and oocyte meiosis, as well as brain size and functioning.[37] There are signals of positive selection, as the result of adaptation to diverse habitats, in genes involved with variation in skin pigmentation and hair morphology.[37] In the immune system, introgressed variants have heavily contributed to the diversity of immune genes, of which there's an enrichment of introgressed alleles that suggest a strong positive selection.[37]
Genes affecting keratin were found to have been introgressed from Neanderthals into modern humans (shown in East Asians and Europeans), suggesting that these genes gave a morphological adaptation in skin and hair to modern humans to cope with non-African environments.[14][18] This is likewise for several genes involved in medical-relevant phenotypes, such as those affecting systemic lupus erythematosus, primary biliary cirrhosis, Crohn's disease, optic disk size, smoking behavior, interleukin 18 levels, and diabetes mellitus type 2.[18]
Researchers found Neanderthal introgression of 18 genes—several of which are related to UV-light adaptation—within the chromosome 3p21.31 region (HYAL region) of East Asians.[38] The introgressive haplotypes were positively selected in only East Asian populations, rising steadily from 45,000 years BP until a sudden increase of growth rate around 5,000 to 3,500 years BP.[38] They occur at very high frequencies among East Asian populations in contrast to other Eurasian populations (e.g. European and South Asian populations).[38] The findings also suggests that this Neanderthal introgression occurred within the ancestral population shared by East Asians and Native Americans.[38]
Evans et al. (2006) had previously suggested that a group of alleles collectively known as haplogroup D of microcephalin, a critical regulatory gene for brain volume, originated from an archaic human population.[39] The results show that haplogroup D introgressed 37,000 years ago (based on the coalescence age of derived D alleles) into modern humans from an archaic human population that separated 1.1 million years ago (based on the separation time between D and non-D alleles), consistent with the period when Neanderthals and modern humans co-existed and diverged respectively.[39] The high frequency of the D haplogroup (70%) suggest that it was positively selected for in modern humans.[39] The distribution of the D allele of microcephalin is high outside Africa but low in sub-Saharan Africa, which further suggest that the admixture event happened in archaic Eurasian populations.[39] This distribution difference between Africa and Eurasia suggests that the D allele originated from Neanderthals according to Lari et al. (2010), but they found that a Neanderthal individual from the Mezzena Rockshelter (Monti Lessini, Italy) was homozygous for an ancestral allele of microcephalin, thus providing no support that Neanderthals contributed the D allele to modern humans and also not excluding the possibility of a Neanderthal origin of the D allele.[40] Green et al. (2010), having analyzed the Vindija Neanderthals, also could not confirm a Neanderthal origin of haplogroup D of the microcephalin gene.[8]
It has been found that HLA-A*02, A*26/*66, B*07, B*51, C*07:02, and C*16:02 of the immune system were contributed from Neanderthals to modern humans.[41] After migrating out of Africa, modern humans encountered and interbred with archaic humans, which was advantageous for modern humans in rapidly restoring HLA diversity and acquiring new HLA variants that are better adapted to local pathogens.[41]
It is found that introgressed Neanderthal genes exhibit cis-regulatory effects in modern humans, contributing to the genomic complexity and phenotype variation of modern humans.[42] Looking at heterozygous individuals (carrying both Neanderthal and modern human versions of a gene), the allele-specific expression of introgressed Neanderthal alleles was found to be significantly lower in the brain and testes relative to other tissues.[4][42] In the brain, this was most pronounced at the cerebellum and basal ganglia.[42] This downregulation suggests that modern humans and Neanderthals possibly experienced a relative higher rate of divergence in these specific tissues.[42]
Furthermore, correlating the genotypes of introgressed Neanderthal alleles with the expression of nearby genes, it is found that archaic alleles contribute proportionally more to variation in expression than nonarchaic alleles.[4] Neanderthal alleles affect expression of the immune genes OAS1/2/3 and TLR1/6/10, which can be specific to cell type and is influenced by environmental stimuli.[4]
Studying the high-coverage female Vindija Neanderthal genome, Prüfer et al. (2017) identified several Neanderthal-derived gene variants, including those that affect levels of LDL cholesterol and vitamin D, and that influence eating disorders, visceral fat accumulation, rheumatoid arthritis, schizophrenia, as well as responses to antipsychotic drugs.[11]
Examining European modern humans in regards to the Altai Neanderthal genome in high-coverage, results show that Neanderthal admixture is associated with several changes in cranium and underlying brain morphology, suggesting changes in neurological function through Neanderthal-derived genetic variation.[43] Neanderthal admixture is associated with an expansion of the posterolateral area of the modern human skull, extending from the occipital and inferior parietal bones to bilateral temporal locales.[43] In regards to modern human brain morphology, Neanderthal admixture is positively correlated with an increase in sulcal depth for the right intraparietal sulcus and an increase in cortical complexity for the early visual cortex of the left hemisphere.[43] Neanderthal admixture is also positively correlated with an increase in white and gray matter volume localized to the right parietal region adjacent to the right intraparietal sulcus.[43] In the area overlapping the primary visual cortex gyrification in the left hemisphere, Neanderthal admixture is positively correlated with gray matter volume.[43] The results also show evidence for a negative correlation between Neanderthal admixture and white matter volume in the orbitofrontal cortex.[43]
In Papuans, assimilated Neanderthal inheritance is found in highest frequency in genes expressed in the brain, whereas Denisovan DNA has the highest frequency in genes expressed in bones and other tissues.[44]
Population substructure theory
Although less parsimonious than recent gene flow, the observation may have been due to ancient population sub-structure in Africa, causing incomplete genetic homogenization within modern humans when Neanderthals diverged while early ancestors of Eurasians were still more closely related to Neanderthals than those of Africans were to Neanderthals.[8] On the basis of allele frequency spectrum, it was shown that the recent admixture model had the best fit to the results while the ancient population sub-structure model had no fit – demonstrating that the best model was a recent admixture event that was preceded by a bottleneck event among modern humans - thus confirming recent admixture as the most parsimonious and plausible explanation for the observed excess of genetic similarities between modern non-African humans and Neanderthals.[45] On the basis of linkage disequilibrium patterns, a recent admixture event is likewise confirmed by the data.[46] From the extent of linkage disequilibrium, it was estimated that the last Neanderthal gene flow into early ancestors of Europeans occurred 47,000–65,000 years BP.[46] In conjunction with archaeological and fossil evidence, the gene flow is thought likely to have occurred somewhere in Western Eurasia, possibly the Middle East.[46] Through another approach—using one genome each of a Neanderthal, Eurasian, African, and chimpanzee (outgroup), and dividing it into non-recombining short sequence blocks—to estimate genome-wide maximum-likelihood under different models, an ancient population sub-structure in Africa was ruled out and a Neanderthal admixture event was confirmed.[10]
Morphology
The early Upper Paleolithic burial remains of a modern human child from Abrigo do Lagar Velho (Portugal) features traits that indicate Neanderthal interbreeding with modern humans dispersing into Iberia.[47] Considering the dating of the burial remains (24,500 years BP) and the persistence of Neanderthal traits long after the transitional period from a Neanderthal to a modern human population in Iberia (28,000–30,000 years BP), the child may have been a descendant of an already heavily admixed population.[47]
The remains of an early Upper Paleolithic modern human from Peștera Muierilor (Romania) of 35,000 years BP shows a morphological pattern of European early modern humans, but possesses archaic or Neanderthal features, suggesting European early modern humans interbreeding with Neanderthals.[48] These features include a large interorbital breadth, a relatively flat superciliary arches, a prominent occipital bun, an asymmetrical and shallow mandibular notch shape, a high mandibular coronoid processus, the relative perpendicular mandibular condyle to notch crest position, and a narrow scapular glenoid fossa.[48]
The early modern human Oase 1 mandible from Peștera cu Oase (Romania) of 34,000–36,000 14C years BP presents a mosaic of modern, archaic, and possible Neanderthal features.[50] It displays a lingual bridging of the mandibular foramen, not present in earlier humans except Neanderthals of the late Middle and Late Pleistocene, thus suggesting affinity with Neanderthals.[50] Concluding from the Oase 1 mandible, there was apparently a significant craniofacial change of early modern humans from at least Europe, possibly due to some degree of admixture with Neanderthals.[50]
The earliest (before about 33 ka BP) European modern humans and the subsequent (Middle Upper Paleolithic) Gravettians, falling anatomically largely in line with the earliest (Middle Paleolithic) African modern humans, also show traits that are distinctively Neanderthal, suggesting that a solely Middle Paleolithic modern human ancestry was unlikely for European early modern humans.[51]
Manot 1, a partial calvarium of a modern human that was recently discovered at the Manot Cave (Western Galilee, Israel) and dated to 54.7±5.5 kyr BP, represents the first fossil evidence from the period when modern humans successfully migrated out of Africa and colonized Eurasia.[52] It also provides the first fossil evidence that modern humans inhabited the southern Levant during the Middle to Upper Palaeolithic interface, contemporaneously with the Neanderthals and close to the probable interbreeding event.[52] The morphological features suggest that the Manot population may be closely related to or may have given rise to the first modern humans who later successfully colonized Europe to establish early Upper Palaeolithic populations.[52]
History
The interbreeding has been discussed ever since the discovery of Neanderthal remains in the 19th century, though earlier writers believed that Neanderthals were a direct ancestor of modern humans. Thomas Huxley suggested that many Europeans bore traces of Neanderthal ancestry, but associated Neanderthal characteristics with primitivism, writing that since they "belong to a stage in the development of the human species, antecedent to the differentiation of any of the existing races, we may expect to find them in the lowest of these races, all over the world, and in the early stages of all races".[53]
Until the early 1950s, most scholars thought Neanderthals were not in the ancestry of living humans.[54]: 232–34 [55] Nevertheless, Hans Peder Steensby proposed interbreeding in 1907 in the article Race studies in Denmark. He strongly emphasised that all living humans are of mixed origins.[56] He held that this would best fit observations, and challenged the widespread idea that Neanderthals were ape-like or inferior. Basing his argument primarily on cranial data, he noted that the Danes, like the Frisians and the Dutch, exhibit some Neanderthaloid characteristics, and felt it was reasonable to "assume something was inherited" and that Neanderthals "are among our ancestors."
Carleton Stevens Coon in 1962 found it likely, based upon evidence from cranial data and material culture, that Neanderthal and Upper Paleolithic peoples either interbred or that the newcomers reworked Neanderthal implements "into their own kind of tools."[57]
By the early 2000s, the majority of scholars supported the Out of Africa hypothesis,[58][59] according to which anatomically modern humans left Africa about 50,000 years ago and replaced Neanderthals with little or no interbreeding. Yet some scholars still argued for hybridisation with Neanderthals. The most vocal proponent of the hybridisation hypothesis was Erik Trinkaus of Washington University.[60] Trinkaus claimed various fossils as products of hybridised populations, including the skeleton of a child found at Lagar Velho in Portugal[61][62][63] and the Peștera Muierii skeletons from Romania.[48]
Denisovans
Genetics

Proportion of admixture
It has been shown that Melanesians (e.g. Papua New Guinean and Bougainville Islander) share relatively more alleles with Denisovans when compared to other Eurasian-derived populations and Africans.[64] It is estimated that 4% to 6% of the genome in Melanesians derives from Denisovans, while no Eurasians or Africans displayed contributions of the Denisovan genes.[64] It has been observed that Denisovans contributed genes to Melanesians but not to East Asians, indicating that there was interaction between the early ancestors of Melanesians with Denisovans but that this interaction did not take place in the regions near southern Siberia, where as-of-yet the only Denisovan remains have been found.[64] In addition, Aboriginal Australians also show a relative increased allele sharing with Denisovans, compared to Eurasians and African populations, consistent with the hypothesis of increased admixture between Denisovans and Melanesians.[65]
Reich et al. (2011) produced evidence that the highest presence of Denisovan admixture is in Oceanian populations, followed by many Southeast Asian populations, and none in East Asian populations.[66] There is significant Denisovan genetic material in eastern Southeast Asian and Oceanian populations (e.g. Aboriginal Australians, Near Oceanians, Polynesians, Fijians, eastern Indonesians, Philippine Mamanwa and Manobo), but not in certain western and continental Southeast Asian populations (e.g. western Indonesians, Malaysian Jehai, Andaman Onge, and mainland Asians), indicating that the Denisovan admixture event happened in Southeast Asia itself rather than mainland Eurasia.[66] The observation of high Denisovan admixture in Oceania and the lack thereof in mainland Asia suggests that early modern humans and Denisovans had interbred east of the Wallace Line that divides Southeast Asia according to Cooper and Stringer (2013).[67]
Skoglund and Jakobsson (2011) observed that particularly Oceanians, followed by Southeast Asians populations, have a high Denisovans admixture relative to other populations.[68] Furthermore, they found possible low traces of Denisovan admixture in East Asians and no Denisovan admixture in Native Americans.[68] In contrast, Prüfer et al. (2013) found that mainland Asian and Native American populations may have a 0.2% Denisovan contribution, which is about twenty-five times lower than Oceanian populations.[9] The manner of gene flow to these populations remains unknown.[9] However, Wall et al. (2013) stated that they found no evidence for Denisovan admixture in East Asians.[17]
Findings indicate that the Denisovan gene flow event happened to the common ancestors of Aboriginal Filipinos, Aboriginal Australians, and New Guineans.[66][69] New Guineans and Australians have similar rates of Denisovan admixture, indicating that interbreeding took place prior to their common ancestors' entry into Sahul (Pleistocene New Guinea and Australia), at least 44,000 years ago.[66] It has also been observed that the fraction of Near Oceanian ancestry in Southeast Asians is proportional to the Denisovan admixture, except in the Philippines where there is a higher proportional Denisovan admixture to Near Oceanian ancestry.[66] Reich et al. (2011) suggested a possible model of an early eastward migration wave of modern humans, some who were Philippine/New Guinean/Australian common ancestors that interbred with Denisovans, respectively followed by divergence of the Philippine early ancestors, interbreeding between the New Guinean and Australian early ancestors with a part of the same early-migration population that did not experience Denisovan gene flow, and interbreeding between the Philippine early ancestors with a part of the population from a much-later eastward migration wave (the other part of the migrating population would become East Asians).[66]
Finding components of Denisovan introgression with differing relatedness to the sequenced Denisovan, Browning et al. (2018) suggested that at least two separate episodes of Denisovan admixture has occurred.[70] Specifically, introgression from two distinct Denisovan populations is observed in East Asians (e.g. Japanese and Han Chinese), whereas South Asians (e.g. Telugu and Punjabi) and Oceanians (e.g. Papuans) display introgression from one Denisovan population.[70]
Exploring derived alleles from Denisovans, Sankararaman et al. (2016) estimated that the date of Denisovan admixture was 44,000–54,000 years ago.[5] They also determined that the Denisovan admixture was the greatest in Oceanian populations compared to other populations with observed Denisovan ancestry (i.e. America, Central Asia, East Asia, and South Asia).[5] The researchers also made the surprising finding that South Asian populations display an elevated Denisovan admixture (when compared to other non-Oceanian populations with Denisovan ancestry), albeit the highest estimate (which are found in Sherpas) is still ten times lower than in Papuans.[5] They suggest two possible explanations: There was a single Denisovan introgression event that was followed by dilution to different extents or at least three distinct pulses of Denisovan introgressions must have occurred.[5]
A study in 2021 analyzing archaic ancestry in 118 Philippine ethnic groups discovered an independent admixture event into Philippine Negritos from Denisovans. The Ayta Magbukon in particular were found to possess the highest level of Denisovan ancestry in the world, with ∼30%–40% more than even that found in Australians and Papuans (Australo-Melanesians), suggesting that distinct Islander Denisovan populations existed in the Philippines which admixed with modern humans after their arrival. [71]
It has been shown that Eurasians have some but significantly lesser archaic-derived genetic material that overlaps with Denisovans, stemming from the fact that Denisovans are related to Neanderthals—who contributed to the Eurasian gene pool—rather than from interbreeding of Denisovans with the early ancestors of those Eurasians.[16][64]
The skeletal remains of an early modern human from the Tianyuan cave (near Zhoukoudian, China) of 40,000 years BP showed a Neanderthal contribution within the range of today's Eurasian modern humans, but it had no discernible Denisovan contribution.[72] It is a distant relative to the ancestors of many Asian and Native American populations, but post-dated the divergence between Asians and Europeans.[72] The lack of a Denisovan component in the Tianyuan individual suggests that the genetic contribution had been always scarce in the mainland.[9]
Reduced contribution
There are large genomic regions devoid of Denisovan-derived ancestry, partly explained by infertility of male hybrids, as suggested by the lower proportion of Denisovan-derived ancestry on X chromosomes and in genes that are expressed in the testes of modern humans.[5]
Changes in modern humans
Exploring the immune system's HLA alleles, it has been suggested that HLA-B*73 introgressed from Denisovans into modern humans in western Asia due to the distribution pattern and divergence of HLA-B*73 from other HLA alleles.[41] Even though HLA-B*73 is not present in the sequenced Denisovan genome, HLA-B*73 was shown to be closely associated to the Denisovan-derived HLA-C*15:05 from the linkage disequilibrium.[41] From phylogenetic analysis, however, it has been concluded that it is highly likely that HLA-B*73 was ancestral.[37]
The Denisovan's two HLA-A (A*02 and A*11) and two HLA-C (C*15 and C*12:02) allotypes correspond to common alleles in modern humans, whereas one of the Denisovan's HLA-B allotype corresponds to a rare recombinant allele and the other is absent in modern humans.[41] It is thought that these must have been contributed from Denisovans to modern humans, because it is unlikely to have been preserved independently in both for so long due to HLA alleles' high mutation rate.[41]
Tibetan people received an advantageous EGLN1 and EPAS1 gene variant, associated with hemoglobin concentration and response to hypoxia, for life at high altitudes from the Denisovans.[37] The ancestral variant of EPAS1 upregulates hemoglobin levels to compensate for low oxygen levels—such as at high altitudes—but this also has the maladaption of increasing blood viscosity.[73] The Denisovan-derived variant on the other hand limits this increase of hemoglobin levels, thus resulting in a better altitude adaption.[73] The Denisovan-derived EPAS1 gene variant is common in Tibetans and was positively selected in their ancestors after they colonized the Tibetan plateau.[73]
Archaic African hominins
Rapid decay of fossils in Sub-Saharan African environments makes it currently unfeasible to compare modern human admixture with reference samples of archaic Sub-Saharan African hominins.[4][74]
From three candidate regions with introgression found by searching for unusual patterns of variations (showing deep haplotype divergence, unusual patterns of linkage disequilibrium, and small basal clade size) in 61 non-coding regions from two hunter-gatherer groups (Biaka Pygmies and San who have significant admixture) and one West African agricultural group (Mandinka, who don't have significant admixture), it is concluded that roughly 2% of the genetic material found in the Biaka Pygmies and San was inserted into the human genome approximately 35,000 years ago from archaic hominins that separated from the ancestors of the modern human lineage around 700,000 years ago.[75] A survey for the introgressive haplotypes across many Sub-Saharan populations suggest that this admixture event happened with archaic hominins who once inhabited Central Africa.[75]
Researching high-coverage whole-genome sequences of fifteen Sub-Saharan hunter-gatherer males from three groups—five Pygmies (three Baka, a Bedzan, and a Bakola) from Cameroon, five Hadza from Tanzania, and five Sandawe from Tanzania—there are signs that the ancestors of the hunter-gatherers interbred with one or more archaic human populations,[74] probably over 40,000 years ago.[76] Analysis of putative introgressive haplotypes in the fifteen hunter-gatherer samples suggests that the archaic African population and modern humans diverged around 1.2 to 1.3 million years ago.[74]
According to a study published in 2020, there are indications that 2% to 19% (or about ≃6.6 and ≃7.0%) of the DNA of four West African populations may have come from an unknown archaic hominin which split from the ancestor of humans and Neanderthals between 360 kya to 1.02 mya. However, the study also finds that at least part of this proposed archaic admixture is also present in Eurasians/non-Africans, and that the admixture event or events range from 0 to 124 ka B.P, which includes the period before the Out-of-Africa migration and prior to the African/Eurasian split (thus affecting in part the common ancestors of both Africans and Eurasians/non-Africans).[77][78][79] Another recent study, which discovered substantial amounts of previously undescribed human genetic variation, also found ancestral genetic variation in Africans that predates modern humans and was lost in most non-Africans.[80]
Related studies
In 2019, scientists discovered evidence, based on genetics studies using artificial intelligence (AI), that suggests the existence of an unknown human ancestor species, not Neanderthal or Denisovan, in the genome of modern humans.[81][82]
See also
- Multiregional origin of modern humans
- Neanderthal extinction
- Interbreeding
References
- based on Schlebusch, CM; Malmström, H; Günther, T; Sjödin, P; Coutinho, A; Edlund, H; Munters, AR; Vicente, M; Steyn, M; Soodyall, H; Lombard, M; Jakobsson, M (2017). "Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago". Science. 358 (6363): 652–655. Bibcode:2017Sci...358..652S. doi:10.1126/science.aao6266. PMID 28971970. Fig. 3 (H. sapiens divergence times) and Stringer, C. (2012). "What makes a modern human". Nature. 485 (7396): 33–35. Bibcode:2012Natur.485...33S. doi:10.1038/485033a. PMID 22552077. S2CID 4420496. (archaic admixture).
- Woodward, Aylin (5 January 2020). "A handful of recent discoveries have shattered anthropologists' picture of where humans came from, and when". Business Insider. Retrieved 6 January 2020.
- Price, Michael (31 January 2020). "Africans, too, carry Neanderthal genetic legacy". Science. 367 (6477): 497. Bibcode:2020Sci...367..497P. doi:10.1126/science.367.6477.497. PMID 32001636. S2CID 210982481.
- Wolf, A. B.; Akey, J. M. (2018). "Outstanding questions in the study of archaic hominin admixture". PLOS Genetics. 14 (5): e1007349. doi:10.1371/journal.pgen.1007349. PMC 5978786. PMID 29852022.
- Sankararaman, Sriram; Mallick, Swapan; Patterson, Nick; Reich, David (2016). "The Combined Landscape of Denisovan and Neanderthal Ancestry in Present-Day Humans". Current Biology. 26 (9): 1241–1247. doi:10.1016/j.cub.2016.03.037. PMC 4864120. PMID 27032491.
- Rogers Ackermann, Rebecca; Mackay, Alex; Arnold, Michael L. (2016). "The Hybrid Origin of "Modern" Humans". Evolutionary Biology. 43: 1–11. doi:10.1007/s11692-015-9348-1. S2CID 14329491.
- "Cro-Magnons Conquered Europe, but Left Neanderthals Alone". PLOS Biology. 2 (12): e449. 30 November 2004. doi:10.1371/journal.pbio.0020449. ISSN 1545-7885. PMC 532398.
- Green, R.E.; Krause, J.; Briggs, A.W.; Maricic, T.; Stenzel, U.; Kircher, M.; et al. (2010). "A Draft Sequence of the Neandertal Genome". Science. 328 (5979): 710–22. Bibcode:2010Sci...328..710G. doi:10.1126/science.1188021. PMC 5100745. PMID 20448178.
- Prüfer, K.; Racimo, F.; Patterson, N.; Jay, F.; Sankararaman, S.; Sawyer, S.; et al. (2014) [Online 2013]. "The complete genome sequence of a Neanderthal from the Altai Mountains". Nature. 505 (7481): 43–49. Bibcode:2014Natur.505...43P. doi:10.1038/nature12886. PMC 4031459. PMID 24352235.
- Lohse, K.; Frantz, L.A.F. (2014). "Neandertal Admixture in Eurasia Confirmed by Maximum-Likelihood Analysis of Three Genomes". Genetics. 196 (4): 1241–51. doi:10.1534/genetics.114.162396. PMC 3982695. PMID 24532731.
- Prüfer, K.; de Filippo, C.; Grote, S.; Mafessoni, F.; Korlević, P.; Hajdinjak, M.; et al. (2017). "A high-coverage Neandertal genome from Vindija Cave in Croatia". Science. 358 (6363): 655–58. Bibcode:2017Sci...358..655P. doi:10.1126/science.aao1887. PMC 6185897. PMID 28982794.
- Chen, Lu; Wolf, Aaron B.; Fu, Wenqing; Li, Liming; Akey, Joshua M. (January 2020). "Identifying and Interpreting Apparent Neanderthal Ancestry in African Individuals". Cell. 180 (4): 677–687.e16. doi:10.1016/j.cell.2020.01.012. PMID 32004458. S2CID 210955842.
- Zimmer, Carl (31 January 2020). "Neanderthal Genes Hint at Much Earlier Human Migration From Africa". The New York Times. Retrieved 31 January 2020.
- Vernot, B.; Akey, J.M. (2014). "Resurrecting Surviving Neandertal Lineages from Modern Human Genomes". Science. 343 (6174): 1017–21. Bibcode:2014Sci...343.1017V. doi:10.1126/science.1245938. PMID 24476670. S2CID 23003860.
- Barras, Colin (2017). "Who are you? How the story of human origins is being rewritten". New Scientist.
Most of us alive today carry inside our cells at least some DNA from a species that last saw the light of day tens of thousands of years ago. And we all carry different bits – to the extent that if you could add them all up, Krause says you could reconstitute something like one-third of the Neanderthal genome and 90 per cent of the Denisovan genome.
- Meyer, M.; Kircher, M.; Gansauge, M.T.; Li, H.; Racimo, F.; Mallick, S.; et al. (2012). "A High-Coverage Genome Sequence from an Archaic Denisovan Individual" (PDF). Science. 338 (6104): 222–26. Bibcode:2012Sci...338..222M. doi:10.1126/science.1224344. PMC 3617501. PMID 22936568.
- Wall, J.D.; Yang, M.A.; Jay, F.; Kim, S.K.; Durand, E.Y.; Stevison, L.S.; et al. (2013). "Higher Levels of Neanderthal Ancestry in East Asians than in Europeans". Genetics. 194 (1): 199–209. doi:10.1534/genetics.112.148213. PMC 3632468. PMID 23410836.
- Sankararaman, S.; Mallick, S.; Dannemann, M.; Prüfer, K.; Kelso, J.; Pääbo, S.; et al. (2014). "The genomic landscape of Neanderthal ancestry in present-day humans". Nature. 507 (7492): 354–57. Bibcode:2014Natur.507..354S. doi:10.1038/nature12961. PMC 4072735. PMID 24476815.
- Nielsen, R.; Akey, J.M.; Jakobsson, M.; Pritchard, J.K.; Tishkoff, S.; Willerslev, E. (2017). "Tracing the peopling of the world through genomics". Nature. 541 (7637): 302–10. Bibcode:2017Natur.541..302N. doi:10.1038/nature21347. PMC 5772775. PMID 28102248.
- Vernot, B.; Akey, J.M. (2015). "Complex History of Admixture between Modern Humans and Neandertals". The American Journal of Human Genetics. 96 (3): 448–53. doi:10.1016/j.ajhg.2015.01.006. PMC 4375686. PMID 25683119.
- Kim, B.Y.; Lohmueller, K.E. (2015). "Selection and Reduced Population Size Cannot Explain Higher Amounts of Neandertal Ancestry in East Asian than in European Human Populations". The American Journal of Human Genetics. 96 (3): 454–61. doi:10.1016/j.ajhg.2014.12.029. PMC 4375557. PMID 25683122.
- Sánchez-Quinto, F.; Botigué, L.R.; Civit, S.; Arenas, C.; Ávila-Arcos, M.C.; Bustamante, C.D.; et al. (2012). "North African Populations Carry the Signature of Admixture with Neandertals". PLOS ONE. 7 (10): e47765. Bibcode:2012PLoSO...747765S. doi:10.1371/journal.pone.0047765. PMC 3474783. PMID 23082212.
We show that North African populations, like all non-African humans, also carry the signature of admixture with Neandertals, and that the real geographical limit for Neandertal admixture is between sub-Saharan groups and the rest[...] our results show that Neandertal genomic traces do not mark a division between African and non-Africans but rather a division between Sub-Saharan Africans and the rest of modern human groups, including those from North Africa.
- Sánchez-Quinto, F.; Botigué, L.R.; Civit, S.; Arenas, C.; Ávila-Arcos, M.C.; Bustamante, C.D.; et al. (2012). "North African Populations Carry the Signature of Admixture with Neandertals". PLOS ONE. 7 (10): e47765. Bibcode:2012PLoSO...747765S. doi:10.1371/journal.pone.0047765. PMC 3474783. PMID 23082212.
We found that North African populations have a significant excess of derived alleles shared with Neandertals, when compared to sub-Saharan Africans. This excess is similar to that found in non-African humans, a fact that can be interpreted as a sign of Neandertal admixture. Furthermore, the Neandertal's genetic signal is higher in populations with a local, pre-Neolithic North African ancestry. Therefore, the detected ancient admixture is not due to recent Near Eastern or European migrations. Sub-Saharan populations are the only ones not affected by the admixture event with Neandertals.
- Sánchez-Quinto, F.; Botigué, L.R.; Civit, S.; Arenas, C.; Ávila-Arcos, M.C.; Bustamante, C.D.; et al. (2012). "North African Populations Carry the Signature of Admixture with Neandertals". PLOS ONE. 7 (10): e47765. Bibcode:2012PLoSO...747765S. doi:10.1371/journal.pone.0047765. PMC 3474783. PMID 23082212.
North African populations have a complex genetic background. In addition to an autochthonous genetic component, they exhibit signals of European, sub-Saharan and Near Eastern admixture as previously described[...] Tunisian Berbers and Saharawi are those populations with highest autochthonous North African component[...] The results of the f4 ancestry ratio test (Table 2 and Table S1) show that North African populations vary in the percentage of Neandertal inferred admixture, primarily depending on the amount of European or Near Eastern ancestry they present (Table 1). Populations like North Morocco and Egypt, with the highest European and Near Eastern component (∼40%), have also the highest amount of Neandertal ancestry (∼60–70%) (Figure 3). On the contrary, South Morocco that exhibits the highest Sub-Saharan component (∼60%), shows the lowest Neandertal signal (20%). Interestingly, the analysis of the Tunisian and N-TUN populations shows a higher Neandertal ancestry component than any other North African population and at least the same (or even higher) as other Eurasian populations (100–138%) (Figure 3).
- Sánchez-Quinto, F.; Botigué, L.R.; Civit, S.; Arenas, C.; Ávila-Arcos, M.C.; Bustamante, C.D.; et al. (2012). "North African Populations Carry the Signature of Admixture with Neandertals". PLOS ONE. 7 (10): e47765. Bibcode:2012PLoSO...747765S. doi:10.1371/journal.pone.0047765. PMC 3474783. PMID 23082212.
Furthermore, the Neandertal's genetic signal is higher in populations with a local, pre-Neolithic North African ancestry. Therefore, the detected ancient admixture is not due to recent Near Eastern or European migrations.
- Wall, J.D.; Yang, M.A.; Jay, F.; Kim, S.K.; Durand, E.Y.; Stevison, L.S.; et al. (2013). "Higher Levels of Neanderthal Ancestry in East Asians than in Europeans". Genetics. 194 (1): 199–209. doi:10.1534/genetics.112.148213. PMC 3632468. PMID 23410836.
Furthermore we find that the Maasai of East Africa have a small but significant fraction of Neanderthal DNA.
- Bekker, Henk (23 October 2017). "Neues Museum in Berlin 1175".
- Kuhlwilm, M.; Gronau, I.; Hubisz, M.J.; de Filippo, C.; Prado-Martinez, J.; Kircher, M.; et al. (2016). "Ancient gene flow from early modern humans into Eastern Neanderthals". Nature. 530 (7591): 429–33. Bibcode:2016Natur.530..429K. doi:10.1038/nature16544. PMC 4933530. PMID 26886800.
- Krings, M.; Stone, A.; Schmitz, R.W.; Krainitzki, H.; Stoneking, M.; Pääbo, Svante (1997). "Neandertal DNA Sequences and the Origin of Modern Humans". Cell. 90 (1): 19–30. doi:10.1016/S0092-8674(00)80310-4. hdl:11858/00-001M-0000-0025-0960-8. PMID 9230299. S2CID 13581775.
- Serre, D.; Langaney, A.; Chech, M.; Teschler-Nicola, M.; Paunovic, M.; Mennecier, P.; et al. (2004). "No Evidence of Neandertal mtDNA Contribution to Early Modern Humans". PLOS Biology. 2 (3): 313–17. doi:10.1371/journal.pbio.0020057. PMC 368159. PMID 15024415.
- Wall, J.D.; Hammer, M.F. (2006). "Archaic admixture in the human genome". Current Opinion in Genetics & Development. 16 (6): 606–10. doi:10.1016/j.gde.2006.09.006. PMID 17027252.
- Mason, P.H.; Short, R.V. (2011). "Neanderthal-human Hybrids". Hypothesis. 9 (1): e1. Archived from the original on 6 December 2019.
- Wang, C.C.; Farina, S.E.; Li, H. (2013) [Online 2012]. "Neanderthal DNA and modern human origins". Quaternary International. 295: 126–29. Bibcode:2013QuInt.295..126W. doi:10.1016/j.quaint.2012.02.027.
- Petr, Martin; Hajdinjak, Mateja; Fu, Qiaomei; Essel, Elena; Rougier, Hélène; Crevecoeur, Isabelle; et al. (25 September 2020). "The evolutionary history of Neanderthal and Denisovan Y chromosomes". Science. 369 (6511): 1653–1656. Bibcode:2020Sci...369.1653P. doi:10.1126/science.abb6460. hdl:21.11116/0000-0007-11C2-A. PMID 32973032. S2CID 221882937.
- Neves, Armando; Serva, Maurizio (2012). "Extremely Rare Interbreeding Events Can Explain Neanderthal DNA in Living Humans". PLOS ONE. 7 (10): e47076. Bibcode:2012PLoSO...747076N. doi:10.1371/journal.pone.0047076. PMC 3480414. PMID 23112810.
- Yang, M.A.; Gao, X.; Theunert, C.; Tong, H.; Aximu-Petri, A.; Nickel, B.; et al. (2017). "40,000-Year-Old Individual from Asia Provides Insight into Early Population Structure in Eurasia". Current Biology. 27 (20): 3202–08.e9. doi:10.1016/j.cub.2017.09.030. PMC 6592271. PMID 29033327.
- Dolgova, O.; Lao, O. (18 July 2018). "Evolutionary and Medical Consequences of Archaic Introgression into Modern Human Genomes". Genes. 9 (7): 358. doi:10.3390/genes9070358. PMC 6070777. PMID 30022013.
- Ding, Q.; Hu, Y.; Xu, S.; Wang, J.; Jin, L. (2014) [Online 2013]. "Neanderthal Introgression at Chromosome 3p21.31 was Under Positive Natural Selection in East Asians". Molecular Biology and Evolution. 31 (3): 683–95. doi:10.1093/molbev/mst260. PMID 24336922.
- Evans, P.D.; Mekel-Bobrov, N.; Vallender, E.J.; Hudson, R.R.; Lahn, B.T. (2006). "Evidence that the adaptive allele of the brain size gene microcephalin introgressed into Homo sapiens from an archaic Homo lineage". Proceedings of the National Academy of Sciences. 103 (48): 18178–83. Bibcode:2006PNAS..10318178E. doi:10.1073/pnas.0606966103. PMC 1635020. PMID 17090677.
- Lari, M.; Rizzi, E.; Milani, L.; Corti, G.; Balsamo, C.; Vai, S.; et al. (2010). "The Microcephalin Ancestral Allele in a Neanderthal Individual". PLOS ONE. 5 (5): e10648. Bibcode:2010PLoSO...510648L. doi:10.1371/journal.pone.0010648. PMC 2871044. PMID 20498832.
- Abi-Rached, L.; Jobin, M. J.; Kulkarni, S.; McWhinnie, A.; Dalva, K.; Gragert, L.; et al. (2011). "The Shaping of Modern Human Immune Systems by Multiregional Admixture with Archaic Humans". Science. 334 (6052): 89–94. Bibcode:2011Sci...334...89A. doi:10.1126/science.1209202. PMC 3677943. PMID 21868630.
- McCoy, R.C.; Wakefield, J.; Akey, J.M. (2017). "Impacts of Neanderthal-Introgressed Sequences on the Landscape of Human Gene Expression". Cell. 168 (5): 916–27. doi:10.1016/j.cell.2017.01.038. PMC 6219754. PMID 28235201.
- Gregory, M.D.; Kippenhan, J.S.; Eisenberg, D.P.; Kohn, P.D.; Dickinson, D.; Mattay, V.S.; et al. (2017). "Neanderthal-Derived Genetic Variation Shapes Modern Human Cranium and Brain". Scientific Reports. 7 (1): 6308. Bibcode:2017NatSR...7.6308G. doi:10.1038/s41598-017-06587-0. PMC 5524936. PMID 28740249.
- Akkuratov, Evgeny E; Gelfand, Mikhail S; Khrameeva, Ekaterina E (2018). "Neanderthal and Denisovan ancestry in Papuans: A functional study". Journal of Bioinformatics and Computational Biology. 16 (2): 1840011. doi:10.1142/S0219720018400115. PMID 29739306.
- Yang, M.A.; Malaspinas, A.S.; Durand, E.Y.; Slatkin, M. (2012). "Ancient Structure in Africa Unlikely to Explain Neanderthal and Non-African Genetic Similarity". Molecular Biology and Evolution. 29 (10): 2987–95. doi:10.1093/molbev/mss117. PMC 3457770. PMID 22513287.
- Sankararaman, S.; Patterson, N.; Li, H.; Pääbo, S.; Reich, D; Akey, J.M. (2012). "The Date of Interbreeding between Neandertals and Modern Humans". PLOS Genetics. 8 (10): e1002947. arXiv:1208.2238. Bibcode:2012arXiv1208.2238S. doi:10.1371/journal.pgen.1002947. PMC 3464203. PMID 23055938.
- Duarte, C.; Maurício, J.; Pettitt, P.B.; Souto, P.; Trinkaus, E.; Plicht, H. van der; Zilhão, J. (1999). "The early Upper Paleolithic human skeleton from the Abrigo do Lagar Velho (Portugal) and modern-human emergence in Iberia". Proceedings of the National Academy of Sciences. 96 (13): 7604–09. Bibcode:1999PNAS...96.7604D. doi:10.1073/pnas.96.13.7604. PMC 22133. PMID 10377462.
- Soficaru, Andrei; Dobos, Adrian; Trinkaus, Erik (2006). "Early modern humans from the Pestera Muierii, Baia de Fier, Romania". Proceedings of the National Academy of Sciences. 103 (46): 17196–201. Bibcode:2006PNAS..10317196S. doi:10.1073/pnas.0608443103. JSTOR 30052409. PMC 1859909. PMID 17085588.
- "Oase 2". Smithsonian National Museum of Natural History. 23 January 2010. Retrieved 1 May 2018.
- Trinkaus E.; Moldovan O.; Milota S.; Bîlgăr A.; Sarcina L.; Athreya S.; et al. (2003). "An early modern human from the Peştera cu Oase, Romania". Proceedings of the National Academy of Sciences. 100 (20): 11231–36. Bibcode:2003PNAS..10011231T. doi:10.1073/pnas.2035108100. PMC 208740. PMID 14504393.
- Trinkaus, E. (2007). "European early modern humans and the fate of the Neandertals". Proceedings of the National Academy of Sciences. 104 (18): 7367–72. Bibcode:2007PNAS..104.7367T. doi:10.1073/pnas.0702214104. PMC 1863481. PMID 17452632.
- Hershkovitz, Israel; Marder, Ofer; Ayalon, Avner; Bar-Matthews, Miryam; Yasur, Gal; Boaretto, Elisabetta; et al. (28 January 2015). "Levantine cranium from Manot Cave (Israel) foreshadows the first European modern humans". Nature. 520 (7546): 216–19. Bibcode:2015Natur.520..216H. doi:10.1038/nature14134. PMID 25629628. S2CID 4386123.
- Huxley, T. (1890). "The Aryan Question and Pre-Historic Man". Collected Essays: Volume VII, Man's Place in Nature.
- Boule, Marcellin (1911–1913). "L'homme fossile de La Chapelle-aux-Saints". Annales de Paléontologie (in French). 6–8.
- G.E. Smith (1928). "Neanderthal Man Not Our Ancestor". Scientific American. 139 (2): 112–15. Bibcode:1928SciAm.139..112S. doi:10.1038/scientificamerican0828-112.(subscription required)
- Steensby, H. P. (1907). "Racestudier i Danmark" [Race Studies in Denmark] (PDF). Geographical Journal (in Danish). Royal Library, Denmark. Retrieved 6 July 2017.
- Coon, Carleton Stevens (1962). "The Origin of races". Science. New York: Knopf. 140 (3563): 208. doi:10.1126/science.140.3563.208. PMID 14022816.
- Liu, Prugnolle et al. (2006). "Currently available genetic and archaeological evidence is supportive of a recent single origin of modern humans in East Africa. However, this is where the consensus on human settlement history ends, and considerable uncertainty clouds any more detailed aspect of human colonization history."
- Stringer, Chris (June 2003). "Human evolution: Out of Ethiopia". Nature. 423 (6941): 692–93, 695. Bibcode:2003Natur.423..692S. doi:10.1038/423692a. PMID 12802315. S2CID 26693109.
- Dan Jones: The Neanderthal within., New Scientist 193.2007, H. 2593 (3 March), 28–32. Modern Humans, Neanderthals May Have Interbred ; Humans and Neanderthals interbred Archived 22 February 2009 at the Wayback Machine
- Foley, Jim (31 July 2000). "The Lagar Velho 1 Skeleton". Fossil Hominids FAQ. TalkOrigins Archive. Retrieved 6 July 2017.
- Sample, Ian (13 September 2006). "Life on the edge: was a Gibraltar cave last outpost of the lost neanderthal?". The Guardian. Retrieved 6 July 2017.
- "Not a lasting last for the Neandertals". john hawks weblog. 13 September 2006. Retrieved 6 July 2017.
- Reich, D.; Green, R.E.; Kircher, M.; Krause, J.; Patterson, N.; Durand, E.Y.; et al. (2010). "Genetic history of an archaic hominin group from Denisova Cave in Siberia" (PDF). Nature. 468 (7327): 1053–60. Bibcode:2010Natur.468.1053R. doi:10.1038/nature09710. hdl:10230/25596. PMC 4306417. PMID 21179161.
- Rasmussen, M.; Guo, X.; Wang, Y.; Lohmueller, K.E.; Rasmussen, S.; Albrechtsen, A.; et al. (2011). "An Aboriginal Australian Genome Reveals Separate Human Dispersals into Asia". Science. 334 (6052): 94–98. Bibcode:2011Sci...334...94R. doi:10.1126/science.1211177. PMC 3991479. PMID 21940856.
- Reich, D.; Patterson, N.; Kircher, M.; Delfin, F.; Nandineni, M.R.; Pugach, I.; et al. (2011). "Denisova Admixture and the First Modern Human Dispersals into Southeast Asia and Oceania". The American Journal of Human Genetics. 89 (4): 516–28. doi:10.1016/j.ajhg.2011.09.005. PMC 3188841. PMID 21944045.
- Cooper, A.; Stringer, C.B. (2013). "Did the Denisovans Cross Wallace's Line?". Science. 342 (6156): 321–23. Bibcode:2013Sci...342..321C. doi:10.1126/science.1244869. PMID 24136958. S2CID 206551893.
- Skoglund, P.; Jakobsson, M. (2011). "Archaic human ancestry in East Asia". Proceedings of the National Academy of Sciences. 108 (45): 18301–06. Bibcode:2011PNAS..10818301S. doi:10.1073/pnas.1108181108. PMC 3215044. PMID 22042846.
- Flatow, I.; Reich, D. (31 August 2012). "Meet Your Ancient Relatives: The Denisovans". NPR.
- Browning, S.R.; Browning, B.L.; Zhou, Y.; Tucci, S.; Akey, J.M. (2018). "Analysis of Human Sequence Data Reveals Two Pulses of Archaic Denisovan Admixture". Cell. 173 (1): 53–61.e9. doi:10.1016/j.cell.2018.02.031. PMC 5866234. PMID 29551270.
- Larena, Maximilian; McKenna, James; Sanchez-Quinto, Federico (2021). "Philippine Ayta possess the highest level of Denisovan ancestry in the world". Current Biology. 31 (19): 4219–4230. doi:10.1016/j.cub.2021.07.022. PMC 8596304. PMID 34388371.
- Fu, Q.; Meyer, M.; Gao, X.; Stenzel, U.; Burbano, H.A.; Kelso, J.; Paabo, S. (2013). "DNA analysis of an early modern human from Tianyuan Cave, China". Proceedings of the National Academy of Sciences. 110 (6): 2223–27. Bibcode:2013PNAS..110.2223F. doi:10.1073/pnas.1221359110. PMC 3568306. PMID 23341637.
- Huerta-Sánchez, E.; Jin, X.; Asan; Bianba, Z.; Peter, B.M.; Vinckenbosch, N.; et al. (2014). "Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA". Nature. 512 (7513): 194–97. Bibcode:2014Natur.512..194H. doi:10.1038/nature13408. PMC 4134395. PMID 25043035.
- Lachance, J.; Vernot, B.; Elbers, C.C.; Ferwerda, B.; Froment, A.; Bodo, J.M.; et al. (2012). "Evolutionary History and Adaptation from High-Coverage Whole-Genome Sequences of Diverse African Hunter-Gatherers". Cell. 150 (3): 457–69. doi:10.1016/j.cell.2012.07.009. PMC 3426505. PMID 22840920.
- Hammer, M.F.; Woerner, A.E.; Mendez, F.L.; Watkins, J.C.; Wall, J.D. (2011). "Genetic evidence for archaic admixture in Africa". Proceedings of the National Academy of Sciences. 108 (37): 15123–28. Bibcode:2011PNAS..10815123H. doi:10.1073/pnas.1109300108. PMC 3174671. PMID 21896735.
- Callaway, E. (2012). "Hunter-gatherer genomes a trove of genetic diversity". Nature. doi:10.1038/nature.2012.11076. S2CID 87081207.
- Arun Durvasula; Sriram Sankararaman (2020). "Recovering signals of ghost archaic introgression in African populations". Science Advances. 6 (7): eaax5097. Bibcode:2020SciA....6.5097D. doi:10.1126/sciadv.aax5097. PMC 7015685. PMID 32095519. "Non-African populations (Han Chinese in Beijing and Utah residents with northern and western European ancestry) also show analogous patterns in the CSFS, suggesting that a component of archaic ancestry was shared before the split of African and non-African populations...One interpretation of the recent time of introgression that we document is that archaic forms persisted in Africa until fairly recently. Alternately, the archaic population could have introgressed earlier into a modern human population, which then subsequently interbred with the ancestors of the populations that we have analyzed here. The models that we have explored here are not mutually exclusive, and it is plausible that the history of African populations includes genetic contributions from multiple divergent populations, as evidenced by the large effective population size associated with the introgressing archaic population...Given the uncertainty in our estimates of the time of introgression, we wondered whether jointly analyzing the CSFS from both the CEU (Utah residents with Northern and Western European ancestry) and YRI genomes could provide additional resolution. Under model C, we simulated introgression before and after the split between African and non-African populations and observed qualitative differences between the two models in the high-frequency–derived allele bins of the CSFS in African and non-African populations (fig. S40). Using ABC to jointly fit the high-frequency–derived allele bins of the CSFS in CEU and YRI (defined as greater than 50% frequency), we find that the lower limit on the 95% credible interval of the introgression time is older than the simulated split between CEU and YRI (2800 versus 2155 generations B.P.), indicating that at least part of the archaic lineages seen in the YRI are also shared with the CEU..."
- Supplementary Materials for Recovering signals of ghost archaic introgression in African populations", section "S8.2" "We simulated data using the same priors in Section S5.2, but computed the spectrum for both YRI [West African Yoruba] and CEU [a population of European origin] . We found that the best fitting parameters were an archaic split time of 27,000 generations ago (95% HPD: 26,000-28,000), admixture fraction of 0.09 (95% HPD: 0.04-0.17), admixture time of 3,000 generations ago (95% HPD: 2,800-3,400), and an effective population size of 19,700 individuals (95% HPD: 19,300-20,200). We find that the lower bound of the admixture time is further back than the simulated split between CEU and YRI (2155 generations ago), providing some evidence in favor of a pre-Out-of-Africa event. This model suggests that many populations outside of Africa should also contain haplotypes from this introgression event, though detection is difficult because many methods use unadmixed outgroups to detect introgressed haplotypes [Browning et al., 2018, Skov et al., 2018, Durvasula and Sankararaman, 2019] (5, 53, 22). It is also possible that some of these haplotypes were lost during the Out-of-Africa bottleneck."
- Durvasula, Arun; Sankararaman, Sriram (2020). "Recovering signals of ghost archaic introgression in African populations". Science Advances. 6 (7): eaax5097. Bibcode:2020SciA....6.5097D. doi:10.1126/sciadv.aax5097. PMC 7015685. PMID 32095519.
- Bergström, A; McCarthy, S; Hui, R; Almarri, M; Ayub, Q (2020). "Insights into human genetic variation and population history from 929 diverse genomes". Science. 367 (6484): eaay5012. doi:10.1126/science.aay5012. PMC 7115999. PMID 32193295. "An analysis of archaic sequences in modern populations identifies ancestral genetic variation in African populations that likely predates modern humans and has been lost in most non-African populations...We found small amounts of Neanderthal ancestry in West African genomes, most likely reflecting Eurasian admixture. Despite their very low levels or absence of archaic ancestry, African populations share many Neanderthal and Denisovan variants that are absent from Eurasia, reflecting how a larger proportion of the ancestral human variation has been maintained in Africa."
- Mondal, Mayukh; Bertranpedt, Jaume; Leo, Oscar (16 January 2019). "Approximate Bayesian computation with deep learning supports a third archaic introgression in Asia and Oceania". Nature Communications. 10 (246): 246. Bibcode:2019NatCo..10..246M. doi:10.1038/s41467-018-08089-7. PMC 6335398. PMID 30651539.
- Dockrill, Peter (11 February 2019). "Artificial Intelligence Has Found an Unknown 'Ghost' Ancestor in The Human Genome". ScienceAlert.com. Retrieved 11 February 2019.
