Drosophila melanogaster
Drosophila melanogaster is a species of fly (the taxonomic order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly" or "pomace fly".[lower-alpha 1][4] Starting with Charles W. Woodworth's 1901 proposal of the use of this species as a model organism,[5][6] D. melanogaster continues to be widely used for biological research in genetics, physiology, microbial pathogenesis, and life history evolution. As of 2017, five Nobel Prizes have been awarded to drosophilists for their work using the insect.[7][8]
| Drosophila melanogaster | |
|---|---|
 | |
| Scientific classification | |
| Kingdom: | Animalia |
| Phylum: | Arthropoda |
| Class: | Insecta |
| Order: | Diptera |
| Family: | Drosophilidae |
| Genus: | Drosophila |
| Subgenus: | Sophophora |
| Species group: | melanogaster |
| Species subgroup: | melanogaster |
| Species complex: | melanogaster |
| Species: | D. melanogaster |
| Binomial name | |
| Drosophila melanogaster Meigen, 1830[1] | |
D. melanogaster is typically used in research owing to its rapid life cycle, relatively simple genetics with only four pairs of chromosomes, and large number of offspring per generation.[9] It was originally an African species, with all non-African lineages having a common origin.[10] Its geographic range includes all continents, including islands.[11] D. melanogaster is a common pest in homes, restaurants, and other places where food is served.[12]
Flies belonging to the family Tephritidae are also called "fruit flies". This can cause confusion, especially in the Mediterranean, Australia, and South Africa, where the Mediterranean fruit fly Ceratitis capitata is an economic pest.
Physical appearance
.jpg.webp)
.jpg.webp)
Wild type fruit flies are yellow-brown, with brick-red eyes and transverse black rings across the abdomen. The brick-red color of the eyes of the wild type fly are due to two pigments:[13] xanthommatin, which is brown and is derived from tryptophan, and drosopterins, which are red and are derived from guanosine triphosphate.[13] They exhibit sexual dimorphism; females are about 2.5 mm (0.10 in) long; males are slightly smaller with darker backs. Males are easily distinguished from females based on colour differences, with a distinct black patch at the abdomen, less noticeable in recently emerged flies, and the sexcombs (a row of dark bristles on the tarsus of the first leg). Furthermore, males have a cluster of spiky hairs (claspers) surrounding the reproducing parts used to attach to the female during mating. Extensive images are found at FlyBase.[14] Drosophila melanogaster flies can sense air currents with the hairs on their backs. Their eyes are sensitive to slight differences in light intensity and will instinctively fly away when a shadow or other movement is detected.[15]
Lifecycle and reproduction
Under optimal growth conditions at 25 °C (77 °F), the D. melanogaster lifespan is about 50 days from egg to death.[16] The developmental period for D. melanogaster varies with temperature, as with many ectothermic species. The shortest development time (egg to adult), 7 days, is achieved at 28 °C (82 °F).[17][18] Development times decrease at higher temperatures (11 days at 30 °C or 86 °F) due to heat stress. Under ideal conditions, the development time at 25 °C (77 °F) is 8.5 days,[17][18][19] at 18 °C (64 °F) it takes 19 days[17][18] and at 12 °C (54 °F) it takes over 50 days.[17][18] Under crowded conditions, development time increases,[20] while the emerging flies are smaller.[20][21] Females lay some 400 eggs (embryos), about five at a time, into rotting fruit or other suitable material such as decaying mushrooms and sap fluxes. Drosophila melanogaster is a holometabolous insect, so it undergoes a full metamorphosis. Their life cycle is broken down into 4 stages: embryo, larva, pupa, adult.[22] The eggs, which are about 0.5 mm long, hatch after 12–15 hours (at 25 °C or 77 °F).[17][18] The resulting larvae grow for about 4 days (at 25 °C) while molting twice (into second- and third-instar larvae), at about 24 and 48 h after hatching.[17][18] During this time, they feed on the microorganisms that decompose the fruit, as well as on the sugar of the fruit itself. The mother puts feces on the egg sacs to establish the same microbial composition in the larvae's guts that has worked positively for herself.[23] Then the larvae encapsulate in the puparium and undergo a 4-day-long metamorphosis (at 25 °C), after which the adults eclose (emerge).[17][18]
Males perform a sequence of five behavioral patterns to court females. First, males orient themselves while playing a courtship song by horizontally extending and vibrating their wings. Soon after, the male positions himself at the rear of the female's abdomen in a low posture to tap and lick the female genitalia. Finally, the male curls his abdomen and attempts copulation. Females can reject males by moving away, kicking, and extruding their ovipositor.[24] Copulation lasts around 15–20 minutes,[25] during which males transfer a few hundred, very long (1.76 mm) sperm cells in seminal fluid to the female.[26] Females store the sperm in a tubular receptacle and in two mushroom-shaped spermathecae; sperm from multiple matings compete for fertilization. A last male precedence is believed to exist; the last male to mate with a female sires about 80% of her offspring. This precedence was found to occur through both displacement and incapacitation.[27] The displacement is attributed to sperm handling by the female fly as multiple matings are conducted and is most significant during the first 1–2 days after copulation. Displacement from the seminal receptacle is more significant than displacement from the spermathecae.[27] Incapacitation of first male sperm by second male sperm becomes significant 2–7 days after copulation. The seminal fluid of the second male is believed to be responsible for this incapacitation mechanism (without removal of first male sperm) which takes effect before fertilization occurs.[27] The delay in effectiveness of the incapacitation mechanism is believed to be a protective mechanism that prevents a male fly from incapacitating his own sperm should he mate with the same female fly repetitively. Sensory neurons in the uterus of female D. melanogaster respond to a male protein, sex peptide, which is found in semen.[28] This protein makes the female reluctant to copulate for about 10 days after insemination. The signal pathway leading to this change in behavior has been determined. The signal is sent to a brain region that is a homolog of the hypothalamus and the hypothalamus then controls sexual behavior and desire.[28] Gonadotropic hormones in Drosophila maintain homeostasis and govern reproductive output via a cyclic interrelationship, not unlike the mammalian estrous cycle.[29] Sex peptide perturbs this homeostasis and dramatically shifts the endocrine state of the female by inciting juvenile hormone synthesis in the corpus allatum.[30]
D. melanogaster is often used for life extension studies, such as to identify genes purported to increase lifespan when mutated.[31] D. melanogaster is also used in studies of aging. Werner syndrome is a condition in humans characterized by accelerated aging. It is caused by mutations in the gene WRN that encodes a protein with essential roles in repair of DNA damage. Mutations in the D. melanogaster homolog of WRN also cause increased physiologic signs of aging, such as shorter lifespan, higher tumor incidence, muscle degeneration, reduced climbing ability, altered behavior and reduced locomotor activity.[32]
Females

Females become receptive to courting males about 8–12 hours after emergence.[33] Specific neuron groups in females have been found to affect copulation behavior and mate choice. One such group in the abdominal nerve cord allows the female fly to pause her body movements to copulate.[28] Activation of these neurons induces the female to cease movement and orient herself towards the male to allow for mounting. If the group is inactivated, the female remains in motion and does not copulate. Various chemical signals such as male pheromones often are able to activate the group.[28]
Also, females exhibit mate choice copying. When virgin females are shown other females copulating with a certain type of male, they tend to copulate more with this type of male afterwards than naive females (which have not observed the copulation of others). This behavior is sensitive to environmental conditions, and females copulate less in bad weather conditions.[34]
Males
D. melanogaster males exhibit a strong reproductive learning curve. That is, with sexual experience, these flies tend to modify their future mating behavior in multiple ways. These changes include increased selectivity for courting only intraspecifically, as well as decreased courtship times.
Sexually naïve D. melanogaster males are known to spend significant time courting interspecifically, such as with D. simulans flies. Naïve D. melanogaster will also attempt to court females that are not yet sexually mature, and other males. D. melanogaster males show little to no preference for D. melanogaster females over females of other species or even other male flies. However, after D. simulans or other flies incapable of copulation have rejected the males' advances, D. melanogaster males are much less likely to spend time courting nonspecifically in the future. This apparent learned behavior modification seems to be evolutionarily significant, as it allows the males to avoid investing energy into futile sexual encounters.[35]
In addition, males with previous sexual experience modify their courtship dance when attempting to mate with new females—the experienced males spend less time courting, so have lower mating latencies, meaning that they are able to reproduce more quickly. This decreased mating latency leads to a greater mating efficiency for experienced males over naïve males.[36] This modification also appears to have obvious evolutionary advantages, as increased mating efficiency is extremely important in the eyes of natural selection.
Polygamy
Both male and female D. melanogaster flies act polygamously (having multiple sexual partners at the same time).[37] In both males and females, polygamy results in a decrease in evening activity compared to virgin flies, more so in males than females.[37] Evening activity consists of those in which the flies participate other than mating and finding partners, such as finding food.[38] The reproductive success of males and females varies, because a female only needs to mate once to reach maximum fertility.[38] Mating with multiple partners provides no advantage over mating with one partner, so females exhibit no difference in evening activity between polygamous and monogamous individuals.[38] For males, however, mating with multiple partners increases their reproductive success by increasing the genetic diversity of their offspring.[38] This benefit of genetic diversity is an evolutionary advantage because it increases the chance that some of the offspring will have traits that increase their fitness in their environment.
The difference in evening activity between polygamous and monogamous male flies can be explained with courtship. For polygamous flies, their reproductive success increases by having offspring with multiple partners, and therefore they spend more time and energy on courting multiple females.[38] On the other hand, monogamous flies only court one female, and expend less energy doing so.[38] While it requires more energy for male flies to court multiple females, the overall reproductive benefits it produces has kept polygamy as the preferred sexual choice.[38]
The mechanism that affects courtship behavior in Drosophila is controlled by the oscillator neurons DN1s and LNDs.[39] Oscillation of the DN1 neurons was found to be effected by sociosexual interactions, and is connected to mating-related decrease of evening activity.[39]
Model organism in genetics
D. melanogaster remains one of the most studied organisms in biological research, particularly in genetics and developmental biology. It is also employed in studies of environmental mutagenesis.
History of use in genetic analysis
D. melanogaster was among the first organisms used for genetic analysis, and today it is one of the most widely used and genetically best-known of all eukaryotic organisms. All organisms use common genetic systems; therefore, comprehending processes such as transcription and replication in fruit flies helps in understanding these processes in other eukaryotes, including humans.[40]
Thomas Hunt Morgan began using fruit flies in experimental studies of heredity at Columbia University in 1910 in a laboratory known as the Fly Room. The Fly Room was cramped with eight desks, each occupied by students and their experiments. They started off experiments using milk bottles to rear the fruit flies and handheld lenses for observing their traits. The lenses were later replaced by microscopes, which enhanced their observations. Morgan and his students eventually elucidated many basic principles of heredity, including sex-linked inheritance, epistasis, multiple alleles, and gene mapping.[40]
D. melanogaster had historically been used in laboratories to study genetics and patterns of inheritance. However, D. melanogaster also has importance in environmental mutagenesis research, allowing researchers to study the effects of specific environmental mutagens.[41]
Reasons for use in laboratories

There are many reasons the fruit fly is a popular choice as a model organism:
- Its care and culture require little equipment, space, and expense even when using large cultures.
- It can be safely and readily anesthetized (usually with ether, carbon dioxide gas, by cooling, or with products such as FlyNap).
- Its morphology is easy to identify once anesthetized.
- It has a short generation time (about 10 days at room temperature), so several generations can be studied within a few weeks.
- It has a high fecundity (females lay up to 100 eggs per day, and perhaps 2000 in a lifetime).[9]
- Males and females are readily distinguished, and virgin females can be easily identified by their light-colored, translucent abdomen, facilitating genetic crossing.
- The mature larva has giant chromosomes in the salivary glands called polytene chromosomes, "puffs", which indicate regions of transcription, hence gene activity. The under-replication of rDNA occurs resulting in only 20% of DNA compared to the brain. Compare to the 47%, less rDNA in Sarcophaga barbata ovaries.
- It has only four pairs of chromosomes – three autosomes, and one pair of sex chromosomes.
- Males do not show meiotic recombination, facilitating genetic studies.
- Recessive lethal "balancer chromosomes" carrying visible genetic markers can be used to keep stocks of lethal alleles in a heterozygous state without recombination due to multiple inversions in the balancer.
- The development of this organism—from fertilized egg to mature adult—is well understood.
- Genetic transformation techniques have been available since 1987.
- Its complete genome was sequenced and first published in 2000.[42]
- Sexual mosaics can be readily produced, providing an additional tool for studying the development and behavior of these flies.[43]
Genetic markers

Genetic markers are commonly used in Drosophila research, for example within balancer chromosomes or P-element inserts, and most phenotypes are easily identifiable either with the naked eye or under a microscope. In the list of a few common markers below, the allele symbol is followed by the name of the gene affected and a description of its phenotype. (Note: Recessive alleles are in lower case, while dominant alleles are capitalised.)
- Cy1: Curly; the wings curve away from the body, flight may be somewhat impaired
- e1: Ebony; black body and wings (heterozygotes are also visibly darker than wild type)
- Sb1: Stubble; bristles are shorter and thicker than wild type
- w1: White; eyes lack pigmentation and appear white
- bw: Brown; eye color determined by various pigments combined.
- y1: Yellow; body pigmentation and wings appear yellow, the fly analog of albinism
Classic genetic mutations
Drosophila genes are traditionally named after the phenotype they cause when mutated. For example, the absence of a particular gene in Drosophila will result in a mutant embryo that does not develop a heart. Scientists have thus called this gene tinman, named after the Oz character of the same name.[45] Likewise changes in the Shavenbaby gene cause the loss of dorsal cuticular hairs in Drosophila sechellia larvae.[46] This system of nomenclature results in a wider range of gene names than in other organisms.
- Adh: Alcohol dehydrogenase- Drosophila melanogaster can express the alcohol dehydrogenase (ADH) mutation, thereby preventing the breakdown of toxic levels of alcohols into aldehydes and ketones.[47] While ethanol produced by decaying fruit is a natural food source and location for oviposit for Drosophila at low concentrations (<4%), high concentrations of ethanol can induce oxidative stress and alcohol intoxication.[48] Drosophila's fitness is elevated by consuming the low concentration of ethanol. Initial exposure to ethanol causes hyperactivity, followed by incoordination and sedation.[49] Further research has shown that the antioxidant alpha-ketoglutarate may be beneficial in reducing the oxidative stress produced by alcohol consumption. A 2016 study concluded that food supplementation with 10-mM alpha-ketoglutarate decreased Drosophila alcohol sensitivity over time.[50] For the gene that codes for ADH, there are 194 known classic and insertion alleles.[51] Two alleles that are commonly used for experimentation involving ethanol toxicity and response are ADHs (slow) and ADHF (fast). Numerous experiments have concluded that the two alleles account for the differences in enzymatic activity for each. In comparing Adh-F homozygotes (wild-type) and Adh- nulls (homozygous null), research has shown that Adh- nulls have a lower level of tolerance for ethanol, starting the process of intoxication earlier than its counter partner.[49] Other experiments have also concluded that the Adh allele is haplosufficient. Haplosuffiency states that having one functioning allele will be adequate in producing the needed phenotypes for survival. Meaning that flies that were heterozygous for the Adh allele (one copy of the Adh null allele and one copy of the Adh Wild type allele) gave very similar phenotypical alcohol tolerance as the homozygous dominant flies (two copies of the wild type Adh allele).[48] Regardless of genotype, Drosophila show a negative response to exposure to samples with an ethanol content above 5%, which render any tolerance inadequate, resulting in a lethal dosage and a mortality rate of around 70%.[52] Drosophila show many of the same ethanol responses as humans do. Low doses of ethanol produce hyperactivity, moderate doses incoordination, and high doses sedation.[53]
- b: black- The black mutation was discovered in 1910 by Thomas Hunt Morgan.[54] The black mutation results in a darker colored body, wings, veins, and segments of the fruit fly's leg.[55] This occurs due to the fly's inability to create beta-alanine, a beta amino acid.[54] The phenotypic expression of this mutation varies based on the genotype of the individual; for example, whether the specimen is homozygotic or heterozygotic results in a darker or less dark appearance.[55] This genetic mutation is x-linked recessive.[56]
- bw: brown- The brown eye mutation results from inability to produce or synthesize pteridine (red) pigments, due to a point mutation on chromosome II.[57] When the mutation is homozygous, the pteridine pigments are unable to be synthesized because in the beginning of the pteridine pathway, a defective enzyme is being coded by homozygous recessive genes.[58] In all, mutations in the pteridine pathway produces a darker eye color, hence the resulting color of the biochemical defect in the pteridine pathway being brown.
- m: miniature- One of the first records of the miniature mutation of wings was also made by Thomas Hunt Morgan in 1911. He described the wings as having a similar shape as the wild-type phenotype. However, their miniature designation refers to the lengths of their wings, which do not stretch beyond their body and, thus, are notably shorter than the wild-type length. He also noted its inheritance is connected to the sex of the fly and could be paired with the inheritance of other sex-determined traits such as white eyes.[59] The wings may also demonstrate other characteristics deviant from the wild-type wing, such as a duller and cloudier color.[60] Miniature wings are 1.5x shorter than wild-type but are believed to have the same number of cells. This is due to the lack of complete flattening by these cells, making the overall structure of the wing seem shorter in comparison. The pathway of wing expansion is regulated by a signal-receptor pathway, where the neurohormone bursicon interacts with its complementary G protein-coupled receptor; this receptor drives one of the G-protein subunits to signal further enzyme activity and results in development in the wing, such as apoptosis and growth.[61]
- se: sepia- The eye color of the sepia mutant is sepia, a reddish-brown color. In wild-type flies, ommochromes (brown) and drosopterins (red) give the eyes the typical red color.[62][63] The drosopterins are made via a pathway that involves a pyrimidodiazepine synthase,[64] which is encoded on chromosome 3L. The gene has a premature stop codon in sepia flies, so that the flies cannot produce the pyrimidodiazepine synthase and thus no red pigment, so that the eyes stay sepia.[62] The sepia allele is recessive and thus offspring from sepia flies and homozygous wild type flies, has red eyes. The sepia phenotype does not depend on the sex of the fly.[65]
- v: vermilion- The vermilion mutants cannot produce the brown ommochromes leaving the red drosopterins so that the eyes are vermilion colored (a radiant red) compared to a wild-type D. melanogaster. The vermilion mutation is sex-linked and recessive. The gene that is defect lies on the X chromosome.[66] The brown ommochromes are synthesised from kynurenine, which is made from tryptophane. Vermilion flies cannot convert tryptophane into kynurenine and thus cannot make ommochromes, either.[66] Vermilion mutants live longer than wild-type flies. This longer life span may be associated with the reduced amount of tryptophan converted to kynurenine in vermilion flies.[67]

- vg: vestigial- A spontaneous mutation, discovered in 1919 by Thomas Morgan and Calvin Bridges. Vestigial wings are those not fully developed and that have lost function. Since the discovery of the vestigial gene in Drosophila melanogaster, there have been many discoveries of the vestigial gene in other vertebrates and their functions within the vertebrates.[68] The vestigial gene is considered to be one of the most important genes for wing formation, but when it becomes over expressed the issue of ectopic wings begin to form.[69] The vestigial gene acts to regulate the expression of the wing imaginal discs in the embryo and acts with other genes to regulate the development of the wings. A mutated vestigial allele removes an essential sequence of the DNA required for correct development of the wings.[70]
- w: white- Drosophila melanogaster wild type typically expresses a brick red eye color. The white eye mutation in fruit flies is caused due to the absence of two pigments associated with red and brown eye colors; peridines (red) and ommochromes (brown).[63] In January 1910, Thomas Hunt Morgan first discovered the white gene and denoted it as w. The discovery of the white-eye mutation by Morgan brought about the beginnings of genetic experimentation and analysis of Drosophila melanogaster. Hunt eventually discovered that the gene followed a similar pattern of inheritance related to the meiotic segregation of the X chromosome. He discovered that the gene was located on the X chromosome with this information. This led to the discovery of sex-linked genes and also to the discovery of other mutations in Drosophila melanogaster.[71] The white-eye mutation leads to several disadvantages in flies, such as a reduced climbing ability, shortened life span, and lowered resistance to stress when compared to wild type flies.[72] Drosophila melanogaster has a series of mating behaviors that enable them to copulate within a given environment and therefore contribute to their fitness. After Morgan's discovery of the white-eye mutation being sex-linked, a study led by Sturtevant (1915) concluded that white-eyed males were less successful than wild-type males in terms of mating with females.[73] It was found that the greater the density in eye pigmentation, the greater the success in mating for the males of Drosophila melanogaster.[73]
- y: yellow- The yellow gene is a genetic mutation known as Dmel\y within the widely used data base called FlyBase. This mutation can be easily identified by the atypical yellow pigment observed in the cuticle of the adult flies and the mouth pieces of the larva.[74] The y mutation comprises the following phenotypic classes: the mutants that show a complete loss of pigmentation from the cuticle (y-type) and other mutants that show a mosaic pigment pattern with some regions of the cuticle (wild type, y2-type).[75] The role of the yellow gene is diverse and is responsible for changes in behaviour, sex-specific reproductive maturation and, epigenetic reprogramming.[76] The y gene is an ideal gene to study as it is visibly clear when an organisim has this gene, making it easier to understand the passage of DNA to offspring.[76]

Genome
 D. melanogaster chromosomes to scale with megabase-pair references oriented as in the National Center for Biotechnology Information database, centimorgan distances are approximate and estimated from the locations of selected mapped loci. | |
| NCBI genome ID | 47 |
|---|---|
| Ploidy | diploid |
| Number of chromosomes | 8 |
| Year of completion | 2015 |
The genome of D. melanogaster (sequenced in 2000, and curated at the FlyBase database[42]) contains four pairs of chromosomes – an X/Y pair, and three autosomes labeled 2, 3, and 4. The fourth chromosome is relatively very small and therefore often ignored, aside from its important eyeless gene. The D. melanogaster sequenced genome of 139.5 million base pairs has been annotated[77] and contains around 15,682 genes according to Ensemble release 73. More than 60% of the genome appears to be functional non-protein-coding DNA[78] involved in gene expression control. Determination of sex in Drosophila occurs by the X:A ratio of X chromosomes to autosomes, not because of the presence of a Y chromosome as in human sex determination. Although the Y chromosome is entirely heterochromatic, it contains at least 16 genes, many of which are thought to have male-related functions.[79]
There are three transferrin orthologs, all of which are dramatically divergent from those known in chordate models.[80]
Similarity to humans
A March 2000 study by National Human Genome Research Institute comparing the fruit fly and human genome estimated that about 60% of genes are conserved between the two species.[81] About 75% of known human disease genes have a recognizable match in the genome of fruit flies,[82] and 50% of fly protein sequences have mammalian homologs . An online database called Homophila is available to search for human disease gene homologues in flies and vice versa.[83]
Drosophila is being used as a genetic model for several human diseases including the neurodegenerative disorders Parkinson's, Huntington's, spinocerebellar ataxia and Alzheimer's disease.[84] The fly is also being used to study mechanisms underlying aging and oxidative stress, immunity, diabetes, and cancer, as well as drug abuse.[85][86][87]
Connectome
Drosophila is one of the few animals (C. elegans being another) where detailed neural circuits (a connectome) are available.
A high-level connectome, at the level of brain compartments and interconnecting tracts of neurons, exists for the full fly brain.[88] A version of this is available online.[89]
Detailed circuit-level connectomes exist for the lamina[90][91] and a medulla[92] column, both in the visual system of the fruit fly, and the alpha lobe of the mushroom body.[93]
In May 2017 a paper published in bioRxiv presented an electron microscopy image stack of the whole adult female brain at synaptic resolution. The volume is available for sparse tracing of selected circuits.[94][95]
In 2020, a dense connectome of half the central brain of Drosophila was released,[96] along with a web site that allows queries and exploration of this data.[97] The methods used in reconstruction and initial analysis of the connectome followed.[98]
Development
The life cycle of this insect has four stages: fertilized egg, larva, pupa, and adult.[11]
Embryogenesis in Drosophila has been extensively studied, as its small size, short generation time, and large brood size makes it ideal for genetic studies. It is also unique among model organisms in that cleavage occurs in a syncytium.
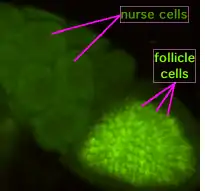
During oogenesis, cytoplasmic bridges called "ring canals" connect the forming oocyte to nurse cells. Nutrients and developmental control molecules move from the nurse cells into the oocyte. In the figure to the left, the forming oocyte can be seen to be covered by follicular support cells.
After fertilization of the oocyte, the early embryo (or syncytial embryo) undergoes rapid DNA replication and 13 nuclear divisions until about 5000 to 6000 nuclei accumulate in the unseparated cytoplasm of the embryo. By the end of the eighth division, most nuclei have migrated to the surface, surrounding the yolk sac (leaving behind only a few nuclei, which will become the yolk nuclei). After the 10th division, the pole cells form at the posterior end of the embryo, segregating the germ line from the syncytium. Finally, after the 13th division, cell membranes slowly invaginate, dividing the syncytium into individual somatic cells. Once this process is completed, gastrulation starts.[99]
Nuclear division in the early Drosophila embryo happens so quickly, no proper checkpoints exist, so mistakes may be made in division of the DNA. To get around this problem, the nuclei that have made a mistake detach from their centrosomes and fall into the centre of the embryo (yolk sac), which will not form part of the fly.
The gene network (transcriptional and protein interactions) governing the early development of the fruit fly embryo is one of the best understood gene networks to date, especially the patterning along the anteroposterior (AP) and dorsoventral (DV) axes (See under morphogenesis).[99]
The embryo undergoes well-characterized morphogenetic movements during gastrulation and early development, including germ-band extension, formation of several furrows, ventral invagination of the mesoderm, and posterior and anterior invagination of endoderm (gut), as well as extensive body segmentation until finally hatching from the surrounding cuticle into a first-instar larva.
During larval development, tissues known as imaginal discs grow inside the larva. Imaginal discs develop to form most structures of the adult body, such as the head, legs, wings, thorax, and genitalia. Cells of the imaginal disks are set aside during embryogenesis and continue to grow and divide during the larval stages—unlike most other cells of the larva, which have differentiated to perform specialized functions and grow without further cell division. At metamorphosis, the larva forms a pupa, inside which the larval tissues are reabsorbed and the imaginal tissues undergo extensive morphogenetic movements to form adult structures.
Developmental plasticity
Biotic and abiotic factors experienced during development will affect developmental resource allocation leading to phenotypic variation, also referred to as developmental plasticity.[100][101] As in all insects,[101] environmental factors can influence several aspects of development in Drosophila melanogaster.[102][103] Fruit flies reared under a hypoxia treatment experience decreased thorax length, while hyperoxia produces smaller flight muscles, suggesting negative developmental effects of extreme oxygen levels.[104] Circadian rhythms are also subject to developmental plasticity. Light conditions during development affect daily activity patterns in Drosophila melanogaster, where flies raised under constant dark or light are less active as adults than those raised under a 12-hour light/dark cycle.[105]
Temperature is one of the most pervasive factors influencing arthropod development. In Drosophila melanogaster temperature-induced developmental plasticity can be beneficial and/or detrimental.[106][107] Most often lower developmental temperatures reduce growth rates which influence many other physiological factors.[108] For example, development at 25 °C increases walking speed, thermal performance breadth, and territorial success, while development at 18 °C increases body mass, wing size, all of which are tied to fitness.[103][106] Moreover, developing at certain low temperatures produces proportionally large wings which improve flight and reproductive performance at similarly low temperatures (See acclimation).[109]
While certain effects of developmental temperature, like body size, are irreversible in ectotherms, others can be reversible.[101][110] When Drosophila melanogaster develop at cold temperatures they will have greater cold tolerance, but if cold-reared flies are maintained at warmer temperatures their cold tolerance decreases and heat tolerance increases over time.[110][111] Because insects typically only mate in a specific range of temperatures, their cold/heat tolerance is an important trait in maximizing reproductive output.[112]
While the traits described above are expected to manifest similarly across sexes, developmental temperature can also produce sex-specific effects in D. melanogaster adults.
- Females- Ovariole number is significantly affected by developmental temperature in D. melanogaster.[113] Egg size is also affected by developmental temperature, and exacerbated when both parents develop at warm temperatures (See Maternal effect).[106] Under stressful temperatures, these structures will develop to smaller ultimate sizes and decrease a female's reproductive output.[113][106] Early fecundity (total eggs laid in first 10 days post-eclosion) is maximized when reared at 25 °C (versus 17 °C and 29 °C) regardless of adult temperature.[114] Across a wide range of developmental temperatures, females tend to have greater heat tolerance than males.[115]
- Males- Stressful developmental temperatures will cause sterility in D. melanogaster males; although the upper temperature limit can be increased by maintaining strains at high temperatures (See acclimation).[107] Male sterility can be reversible if adults are returned to an optimal temperature after developing at stressful temperatures.[116] Male flies are smaller and more successful at defending food/oviposition sites when reared at 25 °C versus 18 °C; thus smaller males will have increased mating success and reproductive output.[103]
Sex determination
Drosophila flies have both X and Y chromosomes, as well as autosomes. Unlike humans, the Y chromosome does not confer maleness; rather, it encodes genes necessary for making sperm. Sex is instead determined by the ratio of X chromosomes to autosomes.[117] Furthermore, each cell "decides" whether to be male or female independently of the rest of the organism, resulting in the occasional occurrence of gynandromorphs.
| X Chromosomes | Autosomes | Ratio of X:A | Sex |
|---|---|---|---|
| XXXX | AAAA | 1 | Normal Female |
| XXX | AAA | 1 | Normal Female |
| XXY | AA | 1 | Normal Female |
| XXYY | AA | 1 | Normal Female |
| XX | AA | 1 | Normal Female |
| XY | AA | 0.50 | Normal Male |
| X | AA | 0.50 | Normal Male (sterile) |
| XXX | AA | 1.50 | Metafemale |
| XXXX | AAA | 1.33 | Metafemale |
| XX | AAA | 0.66 | Intersex |
| X | AAA | 0.33 | Metamale |
Three major genes are involved in determination of Drosophila sex. These are sex-lethal, sisterless, and deadpan. Deadpan is an autosomal gene which inhibits sex-lethal, while sisterless is carried on the X chromosome and inhibits the action of deadpan. An AAX cell has twice as much deadpan as sisterless, so sex-lethal will be inhibited, creating a male. However, an AAXX cell will produce enough sisterless to inhibit the action of deadpan, allowing the sex-lethal gene to be transcribed to create a female.
Later, control by deadpan and sisterless disappears and what becomes important is the form of the sex-lethal gene. A secondary promoter causes transcription in both males and females. Analysis of the cDNA has shown that different forms are expressed in males and females. Sex-lethal has been shown to affect the splicing of its own mRNA. In males, the third exon is included which encodes a stop codon, causing a truncated form to be produced. In the female version, the presence of sex-lethal causes this exon to be missed out; the other seven amino acids are produced as a full peptide chain, again giving a difference between males and females.[118]
Presence or absence of functional sex-lethal proteins now go on to affect the transcription of another protein known as doublesex. In the absence of sex-lethal, doublesex will have the fourth exon removed and be translated up to and including exon 6 (DSX-M[ale]), while in its presence the fourth exon which encodes a stop codon will produce a truncated version of the protein (DSX-F[emale]). DSX-F causes transcription of Yolk proteins 1 and 2 in somatic cells, which will be pumped into the oocyte on its production.
Immunity
The D. melanogaster immune system can be divided into two responses: humoral and cell-mediated. The former is a systemic response mediated in large part through the toll and Imd pathways, which are parallel systems for detecting microbes. Other pathways including the stress response pathways JAK-STAT and P38, nutritional signalling via FOXO, and JNK cell death signalling are all involved in key physiological responses to infection. D. melanogaster has an organ called the "fat body", which is analogous to the human liver. The fat body is the primary secretory organ and produces key immune molecules upon infection, such as serine proteases and antimicrobial peptides (AMPs). AMPs are secreted into the hemolymph and bind infectious bacteria and fungi, killing them by forming pores in their cell walls or inhibiting intracellular processes. The cellular immune response instead refers to the direct activity of blood cells (hemocytes) in Drosophila, which are analogous to mammalian monocytes/macrophages. Hemocytes also possess a significant role in mediating humoral immune responses such as the melanization reaction.[119]
The immune response to infection can involve up to 2,423 genes, or 13.7% of the genome. Although the fly's transcriptional response to microbial challenge is highly specific to individual pathogens, Drosophila differentially expresses a core group of 252 genes upon infection with most bacteria. This core group of genes is associated with gene ontology categories such as antimicrobial response, stress response, secretion, neuron-like, reproduction, and metabolism among others.[120][121] Drosophila also possesses several immune mechanisms to both shape the microbiota and prevent excessive immune responses upon detection of microbial stimuli. For instance, secreted PGRPs with amidase activity scavenge and degrade immunostimulatory DAP-type PGN in order to block Imd activation.[122]
Unlike mammals, Drosophila have innate immunity but lack an adaptive immune response. However, the core elements of this innate immune response are conserved between humans and fruit flies. As a result, the fruit fly offers a useful model of innate immunity for disentangling genetic interactions of signalling and effector function, as flies do not have to contend with interference of adaptive immune mechanisms that could confuse results. Various genetic tools, protocols, and assays make Drosophila a classical model for studying the innate immune system,[123] which has even included immune research on the international space station.[124]
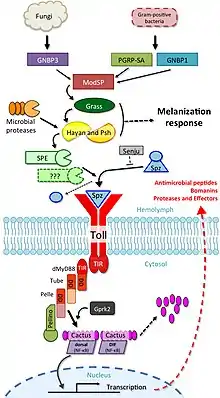
The Drosophila toll pathway
The first description of toll-like receptors involved in the response to infection was performed in Drosophila, culminating in a Nobel prize in 2011.[128][129] The toll pathway in Drosophila is homologous to toll-like pathways in mammals. This regulatory cascade is initiated following pathogen recognition by pattern recognition receptors, particularly of Gram-positive bacteria, parasites, and fungal infection. This activation leads to serine protease signalling cascades ultimately activating the cytokine spätzle. Alternatively, microbial proteases can directly cleave serine proteases like Persephone that then propagate signalling.[130] The cytokine spätzle then acts as the ligand for the toll pathway in flies. Upon infection, pro-spätzle is cleaved by the protease SPE (spätzle-processing enzyme) to become active spätzle, which binds to the toll receptor located on the cell surface of the fat body and dimerizes for activation of downstream NF-κB signaling pathways, including multiple death domain containing proteins and negative regulators such as the ankyrin repeat protein Cactus. The pathway culminates with the translocation of the NF-κB transcription factors Dorsal and Dif (Dorsal-related immunity factor) into the nucleus.[131] Dudzic et al.[132] find a large number of shared serine protease messengers and crosstalk between this pathway and immunity-related melanization pathways.[133][134]
The toll pathway was identified by its regulation of antimicrobial peptides (AMPs), including the antifungal peptide drosomycin. Upon infection, AMPs increase in expression sometimes by 1,000-fold, providing unmistakable readouts of pathway activation. Another group of toll-regulated AMP-like effectors includes the Bomanins, which appear to be responsible for the bulk of toll-mediated immune defence,[135] however Bomanins alone do not exhibit antimicrobial activity.[136]
It has been proposed that a second SPE-like enzyme similarly acts to activate spätzle, as loss of SPE does not completely reduce the activity of toll signalling,[137] however no second SPE has yet been identified. A number of serine proteases are yet to be characterized, including many with homology to SPE.[126] The toll pathway also interacts with renal filtration of microbiota-derived peptidoglycan, maintaining immune homeostasis. Mechanistically, nephrocytes endocytose Lys-type PGN from systemic circulation and route it to lysosomes for degradation. Without this, toll signalling is constitutively activated, resulting in a severe drain on nutrient reserves and a significant stress on host physiology.[138]

The Drosophila Imd pathway
The Imd pathway is orthologous to human TNF receptor superfamily signalling, and is triggered by Gram-negative bacteria through recognition by peptidoglycan recognition proteins (PGRP) including both soluble receptors and cell surface receptors (PGRP-LE and LC, respectively). Imd signalling culminates in the translocation of the NF-κB transcription factor Relish into the nucleus, leading to the upregulation of Imd-responsive genes including the AMP Diptericin. Consequently, flies deficient for AMPs resemble Imd pathway mutants in terms of susceptibility to bacterial infection.[139] Imd signalling and Relish specifically are also involved in the regulation of immunity at surface epithelia including in the gut and respiratory tracts.[119]
The Relish transcription factor has also been implicated in processes regarding cell proliferation[140] and neurodegeneration either through autophagy,[141] or autoimmune toxicity.[142][143] In neurodegenerative models relying on Imd signalling, expression of AMPs in the brain is correlated with brain tissue damage, lesions, and ultimately death.[144][145][146] Relish-regulated AMPs such as Defensin and Diptericin also have anti-cancer properties promoting tumour clearance.[147][148] The Imd-regulated AMP Diptericin B is also produced by the fat body specifically in the head, and Diptericin B is required for long-term memory formation.[149]
JAK-STAT signalling
Multiple elements of the Drosophila JAK-STAT signalling pathway bear direct homology to human JAK-STAT pathway genes. JAK-STAT signalling is induced upon various organismal stresses such as heat stress, dehydration, or infection. JAK-STAT induction leads to the production of a number of stress response proteins including Thioester-containing proteins (TEPs),[150] Turandots,[151] and the putative antimicrobial peptide Listericin.[152] The mechanisms through which many of these proteins act is still under investigation. For instance, the TEPs appear to promote phagocytosis of Gram-positive bacteria and the induction of the toll pathway. As a consequence, flies lacking TEPs are susceptible to infection by toll pathway challenges.[150]
The cellular response to infection
Circulating hemocytes are key regulators of infection. This has been demonstrated both through genetic tools to generate flies lacking hemocytes, or through injecting microglass beads or lipid droplets that saturate hemocyte ability to phagocytose a secondary infection.[153][154] Flies treated like this fail to phagocytose bacteria upon infection, and are correspondingly susceptible to infection.[155] These hemocytes derive from two waves of hematopoiesis, one occurring in the early embryo and one occurring during development from larva to adult.[156] However Drosophila hemocytes do not renew over the adult lifespan, and so the fly has a finite number of hemocytes that decrease over the course of its lifespan.[157] Hemocytes are also involved in regulating cell-cycle events and apoptosis of aberrant tissue (e.g. cancerous cells) by producing Eiger, a tumor necrosis factor signalling molecule that promotes JNK signalling and ultimately cell death and apoptosis.[158]
Behavioral genetics and neuroscience
In 1971, Ron Konopka and Seymour Benzer published "Clock mutants of Drosophila melanogaster", a paper describing the first mutations that affected an animal's behavior. Wild-type flies show an activity rhythm with a frequency of about a day (24 hours). They found mutants with faster and slower rhythms, as well as broken rhythms—flies that move and rest in random spurts. Work over the following 30 years has shown that these mutations (and others like them) affect a group of genes and their products that form a biochemical or biological clock. This clock is found in a wide range of fly cells, but the clock-bearing cells that control activity are several dozen neurons in the fly's central brain.
Since then, Benzer and others have used behavioral screens to isolate genes involved in vision, olfaction, audition, learning/memory, courtship, pain, and other processes, such as longevity.
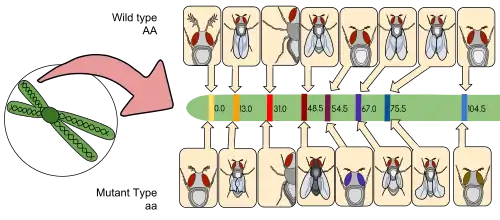
Following the pioneering work of Alfred Henry Sturtevant[159] and others, Benzer and colleagues[43] used sexual mosaics to develop a novel fate mapping technique. This technique made it possible to assign a particular characteristic to a specific anatomical location. For example, this technique showed that male courtship behavior is controlled by the brain.[43] Mosaic fate mapping also provided the first indication of the existence of pheromones in this species.[160] Males distinguish between conspecific males and females and direct persistent courtship preferentially toward females thanks to a female-specific sex pheromone which is mostly produced by the female's tergites.
The first learning and memory mutants (dunce, rutabaga, etc.) were isolated by William "Chip" Quinn while in Benzer's lab, and were eventually shown to encode components of an intracellular signaling pathway involving cyclic AMP, protein kinase A, and a transcription factor known as CREB. These molecules were shown to be also involved in synaptic plasticity in Aplysia and mammals.[161]
The Nobel Prize in Physiology or Medicine for 2017 was awarded to Jeffrey C. Hall, Michael Rosbash, Michael W. Young for their works using fruit flies in understanding the "molecular mechanisms controlling the circadian rhythm".[162]
Male flies sing to the females during courtship using their wings to generate sound, and some of the genetics of sexual behavior have been characterized. In particular, the fruitless gene has several different splice forms, and male flies expressing female splice forms have female-like behavior and vice versa. The TRP channels nompC, nanchung, and inactive are expressed in sound-sensitive Johnston's organ neurons and participate in the transduction of sound.[163][164] Mutating the Genderblind gene, also known as CG6070, alters the sexual behavior of Drosophila, turning the flies bisexual.[165]
Flies use a modified version of Bloom filters to detect novelty of odors, with additional features including similarity of novel odor to that of previously experienced examples, and time elapsed since previous experience of the same odor.[166]
Aggression
As with most insects, aggressive behaviors between male flies commonly occur in the presence of courting a female and when competing for resources. Such behaviors often involve raising wings and legs towards the opponent and attacking with the whole body.[167] Thus, it often causes wing damage, which reduces their fitness by removing their ability to fly and mate.[168]
Acoustic communication
In order for aggression to occur, male flies produce sounds to communicate their intent. A 2017 study found that songs promoting aggression contain pulses occurring at longer intervals.[169] RNA sequencing from fly mutants displaying over-aggressive behaviors found more than 50 auditory-related genes (important for transient receptor potentials, Ca2+ signaling, and mechanoreceptor potentials) to be upregulated in the AB neurons located in Johnston's organ.[169] In addition, aggression levels were reduced when these genes were knocked out via RNA interference.[169] This signifies the major role of hearing as a sensory modality in communicating aggression.
Pheromone signaling
Other than hearing, another sensory modality that regulates aggression is pheromone signaling, which operates through either the olfactory system or the gustatory system depending on the pheromone.[170] An example is cVA, an anti-aphrodisiac pheromone used by males to mark females after copulation and to deter other males from mating.[171] This male-specific pheromone causes an increase in male-male aggression when detected by another male's gustatory system.[170] However, upon inserting a mutation that makes the flies irresponsive to cVA, no aggressive behaviors were seen.[172] This shows how there are multiple modalities for promoting aggression in flies.
Competition for food
Specifically, when competing for food, aggression occurs based on amount of food available and is independent of any social interactions between males.[173] Specifically, sucrose was found to stimulate gustatory receptor neurons, which was necessary to stimulate aggression.[173] However, once the amount of food becomes greater than a certain amount, the competition between males lowers.[173] This is possibly due to an over-abundance of food resources. On a larger scale, food was found to determine the boundaries of a territory since flies were observed to be more aggressive at the food's physical perimeter.
Effect of sleep deprivation
However, like most behaviors requiring arousal and wakefulness, aggression was found to be impaired via sleep deprivation. Specifically, this occurs through the impairment of Octopamine and dopamine signaling, which are important pathways for regulating arousal in insects.[174][175] Due to reduced aggression, sleep-deprived male flies were found to be disadvantaged at mating compared to normal flies.[175] However, when octopamine agonists were administered upon these sleep-deprived flies, aggression levels were seen to be increased and sexual fitness was subsequently restored.[175] Therefore, this finding implicates the importance of sleep in aggression between male flies.
Transgenesis
It is now relatively simple to generate transgenic flies in Drosophila, relying on a variety of techniques. One approach of inserting foreign genes into the Drosophila genome involves P elements. The transposable P elements, also known as transposons, are segments of bacterial DNA that are transferred into the fly genome. Transgenic flies have already contributed to many scientific advances, e.g., modeling such human diseases as Parkinson's, neoplasia, obesity, and diabetes.[176]
Vision

The compound eye of the fruit fly contains 760 unit eyes or ommatidia, and are one of the most advanced among insects. Each ommatidium contains eight photoreceptor cells (R1-8), support cells, pigment cells, and a cornea. Wild-type flies have reddish pigment cells, which serve to absorb excess blue light so the fly is not blinded by ambient light. Eye color genes regulate cellular vesicular transport. The enzymes needed for pigment synthesis are then transported to the cell's pigment granule, which holds pigment precursor molecules.[63]
Each photoreceptor cell consists of two main sections, the cell body and the rhabdomere. The cell body contains the nucleus, while the 100-μm-long rhabdomere is made up of toothbrush-like stacks of membrane called microvilli. Each microvillus is 1–2 μm in length and about 60 nm in diameter.[177] The membrane of the rhabdomere is packed with about 100 million opsin molecules, the visual protein that absorbs light. The other visual proteins are also tightly packed into the microvilli, leaving little room for cytoplasm.
Opsins and spectral sensitivity
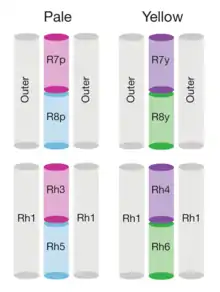
The genome of Drosophila encodes seven opsins,[179] five of those are expressed in the omatidia of the eye. The photoreceptor cells R1-R6 express the opsin Rh1,[180] which absorbs maximally blue light (around 480 nm),[181][182][183] however the R1-R6 cells cover a broader range of the spectrum than an opsin would allow due to a sensitising pigment[184][185] that adds two sensitivity maxima in the UV-range (355 and 370 nm).[183] The R7 cells come in two types with yellow and pale rhabdomeres (R7y and R7p).[186][187] The pale R7p cells express the opsin Rh3,[188][189] which maximally absorbs UV-light (345 nm).[190] The R7p cells are strictly paired with the R8p cells that express Rh5,[189] which maximally absorbs violet light (437 nm).[183] The other, the yellow R7y cells express a blue-absorbing screening pigment[186] and the opsin Rh4,[191] which maximally absorbs UV-light (375 nm).[190] The R7y cells are strictly paired with R8y cells that express Rh6,[192] which maximally absorbs UV-light (508 nm).[183] In a subset of omatidia both R7 and R8 cells express the opsin Rh3.[189]
However, these absorption maxima of the opsins where measured in white eyed flies without screening pigments (Rh3-Rh6),[190][183] or from the isolated opsin directly (Rh1).[181] Those pigments reduce the light that reaches the opsins depending on the wavelength. Thus in fully pigmented flies, the effective absorption maxima of opsins differs and thus also the sensitivity of their photoreceptor cells. With screening pigment, the opsin Rh3 is short wave shifted from 345 nm[lower-alpha 2] to 330 nm and Rh4 from 375 nm to 355 nm. Whether screening pigment is present does not make a practical difference for the opsin Rh5 (435 nm and 437 nm), while the opsin R6 is long wave shifted by 92 nm from 508 nm to 600 nm.[178]
Additionally of the opsins of the eye, Drosophila has two more opsins: The ocelli express the opsin Rh2,[193][194] which maximally absorbs violet light (~420 nm).[194] And the opsin Rh7, which maximally absorbs UV-light (350 nm) with an unusually long wavelength tail up to 500 nm. The long tail disappears if a lysine at position 90 is replaced by glutamic acid. This mutant then absorbs maximally violet light (450 nm).[195] The opsin Rh7 entrains with cryptochrome the circadian rhythm of Drosophila to the day-night-cycle in the central pacemaker neurons.[196]
Each Drosophila opsin binds the carotenoid chromophore 11-cis-3-hydroxyretinal via a lysine.[197][198] This lysine is conserved in almost all opsins, only a few opsins have lost it during evolution.[199] Opsins without it are not light sensitive.[200][201][202] In particular, the Drosophila opsins Rh1, Rh4, and Rh7 function not only as photoreceptors, but also as chemoreceptors for aristolochic acid. These opsins still have the lysine like other opsins. However, if it is replaced by an arginine in Rh1, then Rh1 loses light sensitivity but still responds to aristolochic acid. Thus, the lysine is not needed for Rh1 to function as chemoreceptor.[201]
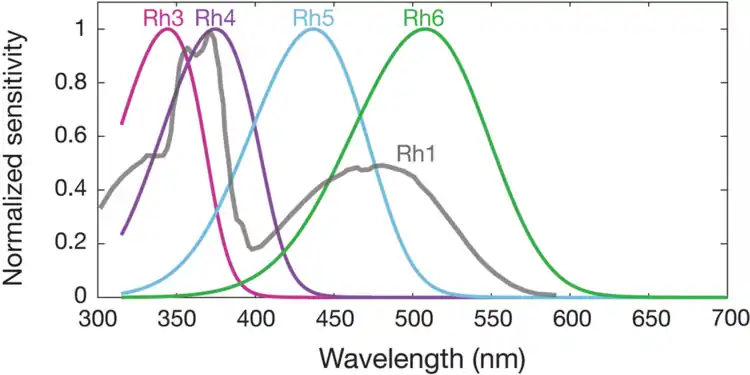 Spectral sensitivities of Drosophila melanogaster opsins in white eyed flies. The sensitivities of Rh3–R6 are modelled with opsin templates and sensitivity estimates from Salcedo et al. (1999).[183] The opsin Rh1 (redrawn from Salcedo et al.[183]) has a characteristic shape as it is coupled to a UV-sensitising pigment.
Spectral sensitivities of Drosophila melanogaster opsins in white eyed flies. The sensitivities of Rh3–R6 are modelled with opsin templates and sensitivity estimates from Salcedo et al. (1999).[183] The opsin Rh1 (redrawn from Salcedo et al.[183]) has a characteristic shape as it is coupled to a UV-sensitising pigment.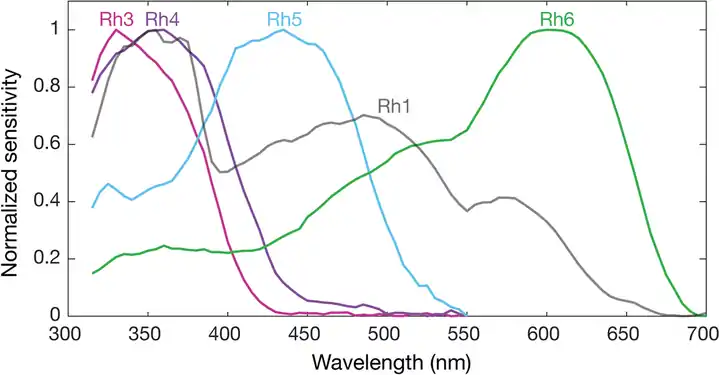 Normalized mean spectral sensitivity curves of Drosophila melanogaster opsins Rh1, Rh3, Rh4, Rh5, and Rh6 measured in their native photoreceptor cells in red eye flies with screening pigment. Each spectral curve is the average from six flies.
Normalized mean spectral sensitivity curves of Drosophila melanogaster opsins Rh1, Rh3, Rh4, Rh5, and Rh6 measured in their native photoreceptor cells in red eye flies with screening pigment. Each spectral curve is the average from six flies.
Phototransduction
As in vertebrate vision, visual transduction in invertebrates occurs via a G protein-coupled pathway. However, in vertebrates, the G protein is transducin, while the G protein in invertebrates is Gq (dgq in Drosophila). When rhodopsin (Rh) absorbs a photon of light its chromophore, 11-cis-3-hydroxyretinal, is isomerized to all-trans-3-hydroxyretinal. Rh undergoes a conformational change into its active form, metarhodopsin. Metarhodopsin activates Gq, which in turn activates a phospholipase Cβ (PLCβ) known as NorpA.[203]
PLCβ hydrolyzes phosphatidylinositol (4,5)-bisphosphate (PIP2), a phospholipid found in the cell membrane, into soluble inositol triphosphate (IP3) and diacylglycerol (DAG), which stays in the cell membrane. DAG or a derivative of DAG causes a calcium-selective ion channel known as transient receptor potential (TRP) to open and calcium and sodium flows into the cell. IP3 is thought to bind to IP3 receptors in the subrhabdomeric cisternae, an extension of the endoplasmic reticulum, and cause release of calcium, but this process does not seem to be essential for normal vision.[203]
Calcium binds to proteins such as calmodulin (CaM) and an eye-specific protein kinase C (PKC) known as InaC. These proteins interact with other proteins and have been shown to be necessary for shut off of the light response. In addition, proteins called arrestins bind metarhodopsin and prevent it from activating more Gq. A sodium-calcium exchanger known as CalX pumps the calcium out of the cell. It uses the inward sodium gradient to export calcium at a stoichiometry of 3 Na+/ 1 Ca++.[204]
TRP, InaC, and PLC form a signaling complex by binding a scaffolding protein called InaD. InaD contains five binding domains called PDZ domain proteins, which specifically bind the C termini of target proteins. Disruption of the complex by mutations in either the PDZ domains or the target proteins reduces the efficiency of signaling. For example, disruption of the interaction between InaC, the protein kinase C, and InaD results in a delay in inactivation of the light response.
Unlike vertebrate metarhodopsin, invertebrate metarhodopsin can be converted back into rhodopsin by absorbing a photon of orange light (580 nm).
About two-thirds of the Drosophila brain is dedicated to visual processing.[205] Although the spatial resolution of their vision is significantly worse than that of humans, their temporal resolution is around 10 times better.
Ceramide kinase is found to increase proapoptotic ceramide activity by Dasgupta et al. 2009, and this increases photoreceptor cell apoptotic turnover.[206]
Grooming
Drosophila are known to exhibit grooming behaviors that are executed in a predictable manner. Drosophila consistently begin a grooming sequence by using their front legs to clean the eyes, then the head and antennae. Using their hind legs, Drosophila proceed to groom their abdomen, and finally the wings and thorax. Throughout this sequence, Drosophila periodically rub their legs together to get rid of excess dust and debris that accumulates during the grooming process.[207]
Grooming behaviors have been shown to be executed in a suppression hierarchy. This means that grooming behaviors that occur at the beginning of the sequence prevent those that come later in the sequence from occurring simultaneously, as the grooming sequence consists of mutually exclusive behaviors.[208][209] This hierarchy does not prevent Drosophila from returning to grooming behaviors that have already been accessed in the grooming sequence.[208] The order of grooming behaviors in the suppression hierarchy is thought to be related to the priority of cleaning a specific body part. For example, the eyes and antennae are likely executed early on in the grooming sequence to prevent debris from interfering with the function of D. melanogaster's sensory organs.[208][209]
Walking

Like many other hexapod insects, Drosophila typically walk using a tripod gait.[211] This means that three of the legs swing together while the other three remain stationary, or in stance. Variability around the tripod configuration appears to be continuous, meaning that flies do not exhibit distinct transitions between different gaits.[212] At fast walking speeds (15–30 mm/s), the walking configuration is mostly tripod (3 legs in stance), but at low walking speeds (0–15 mm/s), flies are more likely to have four or five legs in stance.[213][214] These transitions may help to optimize static stability.[215] Because flies are so small, inertial forces are negligible compared with the elastic forces of their muscles and joints or the viscous forces of the surrounding air.[216]
In addition to stability, the robustness of a walking gait is also thought to be important in determining the gait of a fly at a particular walking speed. Robustness refers to how much offset in the timing of a legs stance can be tolerated before the fly becomes statically unstable.[215] For instance, a robust gait may be particularly important when traversing uneven terrain, as it may cause unexpected disruptions in leg coordination. Using a robust gait would help the fly maintain stability in this case. Analyses suggest that Drosophila may exhibit a compromise between the most stable and most robust gait at a given walking speed.[215]
Flight
Flies fly via straight sequences of movement interspersed by rapid turns called saccades.[217] During these turns, a fly is able to rotate 90° in less than 50 milliseconds.[217]
Characteristics of Drosophila flight may be dominated by the viscosity of the air, rather than the inertia of the fly body, but the opposite case with inertia as the dominant force may occur.[217] However, subsequent work showed that while the viscous effects on the insect body during flight may be negligible, the aerodynamic forces on the wings themselves actually cause fruit flies' turns to be damped viscously.[218]
Misconceptions
Drosophila is sometimes referred to as a pest due to its tendency to live in human settlements, where fermenting fruit is found. Flies may collect in homes, restaurants, stores, and other locations.[12]
The name and behavior of this species of fly has led to the misconception that it is a biological security risk in Australia. While other "fruit fly" species do pose a risk, D. melanogaster is attracted to fruit that is already rotting, rather than causing fruit to rot.[219][220]
See also
- Animal testing on invertebrates
- Eating behavior in Insects (Measurement)
- Fruit flies in space
- Genetically modified insect
- Gynandromorphism
- List of Drosophila databases
- Spätzle (gene)
- Transgenesis
- Zebrafish – another widely used model organism in scientific research
Notes
- "Vinegar fly" is preferred by a handful of recent publications as being a more accurate description than "fruit fly".[2][3][4]
- Sharkey et al.[178] give the absorption maximum of Rh3 as 334 nm in their result section. However, in the introduction and the material and methods section they give it as 345 nm. For both values, they cite Feiler et al., who reported 345 nm only.[190] Therefore, this seems to be a mistake and they probably meant there 345 nm, too.
References
- Meigen JW (1830). Systematische Beschreibung der bekannten europäischen zweiflügeligen Insekten. (Volume 6) (PDF) (in German). Schulz-Wundermann. Archived from the original (PDF) on 2012-02-09.
- "Drosophila | insect genus". Encyclopedia Britannica. Retrieved 2021-10-30.
- "Vinegar Flies". Penn State Extension. Retrieved 2021-10-30.
- Green, M. M. (2002-09-01). "It Really Is Not a Fruit Fly". Genetics. 162 (1): 1–3. doi:10.1093/genetics/162.1.1. ISSN 0016-6731. PMC 1462251. PMID 12242218.
- T.H. Morgan's Nobel Prize biography mentioning C. W. Woodworth
- Holden, Brian (2015-01-01). Charles W. Woodworth: The Remarkable Life of U.C.'s First Entomologist (1 ed.). Brian Holden Publishing. pp. 135–137. ISBN 9780986410536.
- "Nobel Prizes". The Guardian. 7 October 2017.
- "FruitFly-ResearchGate".
- Sang JH (2001-06-23). "Drosophila melanogaster: The Fruit Fly". In Reeve EC (ed.). Encyclopedia of genetics. USA: Fitzroy Dearborn Publishers, I. p. 157. ISBN 978-1-884964-34-3. Retrieved 2009-07-01.
- Baudry E, Viginier B, Veuille M (August 2004). "Non-African populations of Drosophila melanogaster have a unique origin". Molecular Biology and Evolution. 21 (8): 1482–91. doi:10.1093/molbev/msh089. PMID 15014160.
- Markow TA (June 2015). "The secret lives of Drosophila flies". eLife. 4. doi:10.7554/eLife.06793. PMC 4454838. PMID 26041333.
- "Vinegar Flies, Drosophila species, Family: Drosophilidae". Department of Entomology, College of Agricultural Sciences, Pennsylvania State University. 2017. Retrieved 20 July 2017.
- Ewart GD, Howells AJ (1998-01-01). "ABC transporters involved in transport of eye pigment precursors in Drosophila melanogaster". Methods in Enzymology. ABC Transporters: Biochemical, Cellular, and Molecular Aspects. Academic Press. 292: 213–24. doi:10.1016/S0076-6879(98)92017-1. ISBN 9780121821937. PMID 9711556.
- "FlyBase: A database of Drosophila genes and genomes". Genetics Society of America. 2009. Archived from the original on August 15, 2009. Retrieved August 11, 2009.
- "Drosophila Melanogaster". Animal Diversity Web. 2000. Archived from the original on November 30, 2014. Retrieved August 11, 2009.
- Linford NJ, Bilgir C, Ro J, Pletcher SD (January 2013). "Measurement of lifespan in Drosophila melanogaster". Journal of Visualized Experiments (71). doi:10.3791/50068. PMC 3582515. PMID 23328955.
- Ashburner M, Thompson JN (1978). "The laboratory culture of Drosophila". In Ashburner M, Wright TRF (ed.). The genetics and biology of Drosophila. Vol. 2A. Academic Press. 1–81.
- Ashburner M, Golic KG, Hawley RS (2005). Drosophila: A Laboratory Handbook (2nd ed.). Cold Spring Harbor Laboratory Press. pp. 162–4. ISBN 978-0-87969-706-8.
- Bloomington Drosophila Stock Center at Indiana University: Basic Methods of Culturing Drosophila Archived 2006-09-01 at the Wayback Machine
- Chiang HC, Hodson AC (1950). "An analytical study of population growth in Drosophila melanogaster". Ecological Monographs. 20 (3): 173–206. doi:10.2307/1948580. JSTOR 1948580.
- Bakker K (1961). "An analysis of factors which determine success in competition for food among larvae of Drosophila melanogaster". Archives Néerlandaises de Zoologie. 14 (2): 200–281. doi:10.1163/036551661X00061.
- Fernández-Moreno MA, Farr CL, Kaguni LS, Garesse R (2007). "Drosophila melanogaster as a model system to study mitochondrial biology". Mitochondria. Methods in Molecular Biology (Clifton, N.J.). Vol. 372. pp. 33–49. doi:10.1007/978-1-59745-365-3_3. ISBN 978-1-58829-667-2. PMC 4876951. PMID 18314716.
- Blum JE, Fischer CN, Miles J, Handelsman J (November 2013). "Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster". mBio. 4 (6): e00860-13. doi:10.1128/mBio.00860-13. PMC 3892787. PMID 24194543.
- Cook R, Connolly K (1973). "Rejection Responses by Female Drosophila melanogaster: Their Ontogeny, Causality and Effects upon the Behaviour of the Courting Male". Behaviour. 44 (1/2): 142–166. doi:10.1163/156853973x00364. JSTOR 4533484. S2CID 85393769.
- Houot B, Svetec N, Godoy-Herrera R, Ferveur JF (July 2010). "Effect of laboratory acclimation on the variation of reproduction-related characters in Drosophila melanogaster". The Journal of Experimental Biology. 213 (Pt 13): 2322–31. doi:10.1242/jeb.041566. PMID 20543131.
- Gilbert SF (2006). "9: Fertilization in Drosophila". In 8th (ed.). Developmental Biology. Sinauer Associates. ISBN 978-0-87893-250-4. Archived from the original on 2007-02-07.
- Price CS, Dyer KA, Coyne JA (July 1999). "Sperm competition between Drosophila males involves both displacement and incapacitation". Nature. 400 (6743): 449–52. Bibcode:1999Natur.400..449P. doi:10.1038/22755. PMID 10440373. S2CID 4393369.
- "Fruit fly research may reveal what happens in female brains during courtship, mating". Retrieved October 5, 2014.
- Meiselman M, Lee SS, Tran RT, Dai H, Ding Y, Rivera-Perez C, et al. (May 2017). "Drosophila melanogaster". Proceedings of the National Academy of Sciences of the United States of America. 114 (19): E3849–E3858. doi:10.1073/pnas.1620760114. PMC 5441734. PMID 28439025.
- Moshitzky P, Fleischmann I, Chaimov N, Saudan P, Klauser S, Kubli E, Applebaum SW (1996). "Sex-peptide activates juvenile hormone biosynthesis in the Drosophila melanogaster corpus allatum". Archives of Insect Biochemistry and Physiology. 32 (3–4): 363–74. doi:10.1002/(SICI)1520-6327(1996)32:3/4<363::AID-ARCH9>3.0.CO;2-T. PMID 8756302.
- Carnes MU, Campbell T, Huang W, Butler DG, Carbone MA, Duncan LH, et al. (2015). "The Genomic Basis of Postponed Senescence in Drosophila melanogaster". PLOS ONE. 10 (9): e0138569. Bibcode:2015PLoSO..1038569C. doi:10.1371/journal.pone.0138569. PMC 4574564. PMID 26378456.
- Cassidy D, Epiney DG, Salameh C, Zhou LT, Salomon RN, Schirmer AE, et al. (November 2019). "Evidence for premature aging in a Drosophila model of Werner syndrome". Experimental Gerontology. 127: 110733. doi:10.1016/j.exger.2019.110733. PMC 6935377. PMID 31518666.
- Pitnick S (1996). "Investment in testes and the cost of making long sperm in Drosophila". American Naturalist. 148: 57–80. doi:10.1086/285911. S2CID 83654824.
- Dagaeff AC, Pocheville A, Nöbel S, Loyau A, Isabel G, Danchin E (2016). "Drosophila mate copying correlates with atmospheric pressure in a speed learning situation". Animal Behaviour. 121: 163–174. doi:10.1016/j.anbehav.2016.08.022.
- Dukas R (2004). "Male fruit flies learn to avoid interspecific courtship". Behavioral Ecology. 15 (4): 695–698. doi:10.1093/beheco/arh068.
- Saleem S, Ruggles PH, Abbott WK, Carney GE (2014). "Sexual experience enhances Drosophila melanogaster male mating behavior and success". PLOS ONE. 9 (5): e96639. Bibcode:2014PLoSO...996639S. doi:10.1371/journal.pone.0096639. PMC 4013029. PMID 24805129.
- Haartman Lv (1951). "Successive Polygamy". Behaviour. 3 (1): 256–273. doi:10.1163/156853951x00296.
- Vartak VR, Varma V, Sharma VK (February 2015). "Effects of polygamy on the activity/rest rhythm of male fruit flies Drosophila melanogaster". Die Naturwissenschaften. 102 (1–2): 1252. Bibcode:2015SciNa.102....3V. doi:10.1007/s00114-014-1252-5. PMID 25604736. S2CID 7529509.
- Bateman AJ (December 1948). "Intra-sexual selection in Drosophila". Heredity. 2 (Pt. 3): 349–68. doi:10.1038/hdy.1948.21. PMID 18103134.
- Pierce BA (2004). Genetics: A Conceptual Approach (2nd ed.). W. H. Freeman. ISBN 978-0-7167-8881-2.
- Kilbey BJ, MacDonald DJ, Auerbach C, Sobels FH, Vogel EW (June 1981). "The use of Drosophila melanogaster in tests for environmental mutagens". Mutation Research. 85 (3): 141–6. doi:10.1016/0165-1161(81)90029-7. PMID 6790982.
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. (March 2000). "The genome sequence of Drosophila melanogaster". Science. 287 (5461): 2185–95. Bibcode:2000Sci...287.2185.. CiteSeerX 10.1.1.549.8639. doi:10.1126/science.287.5461.2185. PMID 10731132.
- Hotta Y, Benzer S (December 1972). "Mapping of behaviour in Drosophila mosaics". Nature. 240 (5383): 527–35. Bibcode:1972Natur.240..527H. doi:10.1038/240527a0. PMID 4568399. S2CID 4181921.
- Meiers, Sascha (2018). "Exploiting emerging DNA sequencing technologies to study genomic rearrangements". archiv.ub.uni-heidelberg.de. doi:10.11588/heidok.00024506. Retrieved 2021-06-28.
- Azpiazu N, Frasch M (July 1993). "tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila". Genes & Development. 7 (7B): 1325–40. doi:10.1101/gad.7.7b.1325. PMID 8101173.
- Stern DL, Frankel N (December 2013). "The structure and evolution of cis-regulatory regions: the shavenbaby story". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 368 (1632): 20130028. doi:10.1098/rstb.2013.0028. PMC 3826501. PMID 24218640.
- Winberg JO, McKinley-McKee JS (February 1998). "Drosophila melanogaster alcohol dehydrogenase: mechanism of aldehyde oxidation and dismutation". The Biochemical Journal. 329 (Pt 3) (Pt 3): 561–70. doi:10.1042/bj3290561. PMC 1219077. PMID 9445383.
- Ogueta M, Cibik O, Eltrop R, Schneider A, Scholz H (November 2010). "The influence of Adh function on ethanol preference and tolerance in adult Drosophila melanogaster". Chemical Senses. 35 (9): 813–22. doi:10.1093/chemse/bjq084. PMID 20739429.
- Park A, Ghezzi A, Wijesekera TP, Atkinson NS (August 2017). "Genetics and genomics of alcohol responses in Drosophila". Neuropharmacology. 122: 22–35. doi:10.1016/j.neuropharm.2017.01.032. PMC 5479727. PMID 28161376.
- Bayliak MM, Shmihel HV, Lylyk MP, Storey KB, Lushchak VI (September 2016). "Alpha-ketoglutarate reduces ethanol toxicity in Drosophila melanogaster by enhancing alcohol dehydrogenase activity and antioxidant capacity". Alcohol. 55: 23–33. doi:10.1016/j.alcohol.2016.07.009. PMID 27788775.
- "FlyBase Gene Report: Dmel\Adh". flybase.org. Retrieved 2019-03-26.
- Gao HH, Zhai YF, Chen H, Wang YM, Liu Q, Hu QL, Ren FS, Yu Y (September 2018). "Ecological Niche Difference Associated with Varied Ethanol Tolerance between Drosophila suzukii and Drosophila melanogaster (Diptera: Drosophilidae)". Florida Entomologist. 101 (3): 498–504. doi:10.1653/024.101.0308. ISSN 0015-4040.
- Parsch J, Russell JA, Beerman I, Hartl DL, Stephan W (September 2000). "Deletion of a conserved regulatory element in the Drosophila Adh gene leads to increased alcohol dehydrogenase activity but also delays development". Genetics. 156 (1): 219–27. doi:10.1093/genetics/156.1.219. PMC 1461225. PMID 10978287.
- Phillips AM, Smart R, Strauss R, Brembs B, Kelly LE (May 2005). "The Drosophila black enigma: the molecular and behavioural characterization of the black1 mutant allele" (PDF). Gene. 351: 131–42. doi:10.1016/j.gene.2005.03.013. PMID 15878647.
- "FlyBase Gene Report: Dmel\b". flybase.org. Retrieved 2019-03-26.
- Sherald AF (September 1981). "Intergenic suppression of the black mutation of Drosophila melanogaster". Molecular & General Genetics. 183 (1): 102–6. doi:10.1007/bf00270146. PMID 6799739. S2CID 1210971.
- Shoup JR (May 1966). "The development of pigment granules in the eyes of wild type and mutant Drosophila melanogaster". The Journal of Cell Biology. 29 (2): 223–49. doi:10.1083/jcb.29.2.223. PMC 2106902. PMID 5961338.
- "TEACHER REFERENCE PAGES-FLY EYE PIGMENTS LAB" (PDF).
- Morgan TH (March 1911). "The Origin of Nine Wing Mutations in Drosophila". Science. 33 (848): 496–9. Bibcode:1911Sci....33..496M. doi:10.1126/science.33.848.496. JSTOR 1638587. PMID 17774436.
- "FlyBase Gene Report: Dmel\m". flybase.org. Retrieved 2019-03-26.
- Bilousov OO, Katanaev VL, Demydov SV, Kozeretska IA (Mar–Apr 2013). "The downregulation of the miniature gene does not replicate miniature loss-of-function phenotypes in Drosophila melanogaster wing to the full extent". TSitologiia I Genetika. 47 (2): 77–81. PMID 23745366.
- Kim J, Suh H, Kim S, Kim K, Ahn C, Yim J (September 2006). "Identification and characteristics of the structural gene for the Drosophila eye colour mutant sepia, encoding PDA synthase, a member of the omega class glutathione S-transferases". The Biochemical Journal. 398 (3): 451–60. doi:10.1042/BJ20060424. PMC 1559464. PMID 16712527.
- Grant P, Maga T, Loshakov A, Singhal R, Wali A, Nwankwo J, et al. (October 2016). "An Eye on Trafficking Genes: Identification of Four Eye Color Mutations in Drosophila". G3. 6 (10): 3185–3196. doi:10.1534/g3.116.032508. PMC 5068940. PMID 27558665.
- Wiederrecht GJ, Brown GM (1984). "Purification and properties of the enzymes from Drosophila melanogaster that catalyze the conversion of dihydroneopterin triphosphate to the pyrimidodiazepine precursor of the drosopterins". J. Biol. Chem. 259 (22): 14121–7. PMID 6438092.
- "Inheritance Patterns in Drosophila Melanogaster". Retrieved 26 March 2019.
- Green MM (April 1952). "Mutant Isoalleles at the Vermilion Locus in Drosophila Melanogaster". Proceedings of the National Academy of Sciences of the United States of America. 38 (4): 300–5. Bibcode:1952PNAS...38..300G. doi:10.1073/pnas.38.4.300. PMC 1063551. PMID 16589094.
- Oxenkrug, Gregory F. (January 2010). "The extended life span of Drosophila melanogaster eye-color (white and vermilion) mutants with impaired formation of kynurenine". Journal of Neural Transmission. 117 (1): 23–26. doi:10.1007/s00702-009-0341-7. ISSN 0300-9564. PMC 3013506. PMID 19941150.
- Simon E, Faucheux C, Zider A, Thézé N, Thiébaud P (July 2016). "From vestigial to vestigial-like: the Drosophila gene that has taken wing". Development Genes and Evolution. 226 (4): 297–315. doi:10.1007/s00427-016-0546-3. PMID 27116603. S2CID 16651247.
- Tomoyasu Y, Ohde T, Clark-Hachtel C (2017-03-14). "What serial homologs can tell us about the origin of insect wings". F1000Research. 6: 268. doi:10.12688/f1000research.10285.1. PMC 5357031. PMID 28357056.
- Williams JA, Bell JB, Carroll SB (December 1991). "Control of Drosophila wing and haltere development by the nuclear vestigial gene product". Genes & Development. 5 (12B): 2481–95. doi:10.1101/gad.5.12b.2481. PMID 1752439.
- Green MM (January 2010). "2010: A century of Drosophila genetics through the prism of the white gene". Genetics. 184 (1): 3–7. doi:10.1534/genetics.109.110015. PMC 2815926. PMID 20061564.
- Ferreiro MJ, Pérez C, Marchesano M, Ruiz S, Caputi A, Aguilera P, et al. (2018). "rosophila melanogaster White Mutant w1118 Undergo Retinal Degeneration". Frontiers in Neuroscience. 11: 732. doi:10.3389/fnins.2017.00732. PMC 5758589. PMID 29354028.
- Xiao C, Qiu S, Robertson RM (August 2017). "The white gene controls copulation success in Drosophila melanogaster". Scientific Reports. 7 (1): 7712. Bibcode:2017NatSR...7.7712X. doi:10.1038/s41598-017-08155-y. PMC 5550479. PMID 28794482.
- "Gene:Dmel\y". Flybase.org. The FlyBase Consortium. Retrieved 26 March 2019.
- Wittkopp PJ, True JR, Carroll SB (April 2002). "Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns". Development. 129 (8): 1849–58. doi:10.1242/dev.129.8.1849. PMID 11934851.
- Biessmann H (November 1985). "Molecular analysis of the yellow gene (y) region of Drosophila melanogaster". Proceedings of the National Academy of Sciences of the United States of America. 82 (21): 7369–73. Bibcode:1985PNAS...82.7369B. doi:10.1073/pnas.82.21.7369. PMC 391346. PMID 3933004.
- "NCBI (National Center for Biotechnology Information) Genome Database". Retrieved 2011-11-30.
- Halligan DL, Keightley PD (July 2006). "Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison". Genome Research. 16 (7): 875–84. doi:10.1101/gr.5022906. PMC 1484454. PMID 16751341.
- Carvalho AB (December 2002). "Origin and evolution of the Drosophila Y chromosome". Current Opinion in Genetics & Development. 12 (6): 664–8. doi:10.1016/S0959-437X(02)00356-8. PMID 12433579.
- Gabaldón, Toni; Koonin, Eugene V. (2013-04-04). "Functional and evolutionary implications of gene orthology". Nature Reviews Genetics. Nature Portfolio. 14 (5): 360–366. doi:10.1038/nrg3456. ISSN 1471-0056. PMC 5877793. PMID 23552219.
- "Background on Comparative Genomic Analysis". US National Human Genome Research Institute. December 2002.
- Reiter LT, Potocki L, Chien S, Gribskov M, Bier E (June 2001). "A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster". Genome Research. 11 (6): 1114–25. doi:10.1101/gr.169101. PMC 311089. PMID 11381037.
- Chien S, Reiter LT, Bier E, Gribskov M (January 2002). "Homophila: human disease gene cognates in Drosophila". Nucleic Acids Research. 30 (1): 149–51. doi:10.1093/nar/30.1.149. PMC 99119. PMID 11752278.
- Jaiswal M, Sandoval H, Zhang K, Bayat V, Bellen HJ (2012). "Probing mechanisms that underlie human neurodegenerative diseases in Drosophila". Annual Review of Genetics. 46: 371–96. doi:10.1146/annurev-genet-110711-155456. PMC 3663445. PMID 22974305.
- Pick L (2017). Fly Models of Human Diseases. Volume 121 of Current Topics in Developmental Biology. Academic Press. ISBN 978-0-12-802905-3.
- Buchon N, Silverman N, Cherry S (December 2014). "Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology". Nature Reviews. Immunology. 14 (12): 796–810. doi:10.1038/nri3763. PMC 6190593. PMID 25421701.
- Kaun KR, Devineni AV, Heberlein U (June 2012). "Drosophila melanogaster as a model to study drug addiction". Human Genetics. 131 (6): 959–75. doi:10.1007/s00439-012-1146-6. PMC 3351628. PMID 22350798.
- Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, et al. (January 2011). "Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution". Current Biology. 21 (1): 1–11. doi:10.1016/j.cub.2010.11.056. PMID 21129968. S2CID 17155338.
- "FlyCircuit - A Database of Drosophila Brain Neurons". Retrieved 30 Aug 2013.
- Meinertzhagen IA, O'Neil SD (March 1991). "Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster". The Journal of Comparative Neurology. 305 (2): 232–63. doi:10.1002/cne.903050206. PMID 1902848. S2CID 35301798.
- Rivera-Alba M, Vitaladevuni SN, Mishchenko Y, Mischenko Y, Lu Z, Takemura SY, et al. (December 2011). "Wiring economy and volume exclusion determine neuronal placement in the Drosophila brain". Current Biology. 21 (23): 2000–5. doi:10.1016/j.cub.2011.10.022. PMC 3244492. PMID 22119527.
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, et al. (August 2013). "A visual motion detection circuit suggested by Drosophila connectomics". Nature. 500 (7461): 175–81. Bibcode:2013Natur.500..175T. doi:10.1038/nature12450. PMC 3799980. PMID 23925240.
- Takemura SY, Aso Y, Hige T, Wong A, Lu Z, Xu CS, et al. (July 2017). "Drosophila brain". eLife. 6: e26975. doi:10.7554/eLife.26975. PMC 5550281. PMID 28718765.
- "Entire Fruit Fly Brain Imaged with Electron Microscopy". The Scientist Magazine. Retrieved 2018-07-15.
- Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, et al. (July 2018). "A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster". Cell. 174 (3): 730–743.e22. bioRxiv 10.1101/140905. doi:10.1016/j.cell.2018.06.019. PMC 6063995. PMID 30033368.
- Xu CS, Januszewski M, Lu Z, Takemura SY, Hayworth K, Huang G, Shinomiya K, Maitin-Shepard J, Ackerman D, Berg S, Blakely T, et al. (2020). "A connectome of the adult Drosophila central brain". bioRxiv. Cold Spring Harbor Laboratory: 2020.01.21.911859. doi:10.1101/2020.01.21.911859. S2CID 213140797.
- "Analysis tools for connectomics". HHMI.
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura SY, Hayworth KJ, Huang G, Shinomiya K, Maitlin-Shepard J, Berg S, Clements J, et al. (2020). "A Connectome and Analysis of the Adult Drosophila Central Brain". bioRxiv. Cold Spring Harbor. 9. doi:10.1101/2020.04.07.030213. PMC 7546738. PMID 32880371. S2CID 215790785.
- Weigmann K, Klapper R, Strasser T, Rickert C, Technau G, Jäckle H, et al. (June 2003). "FlyMove--a new way to look at development of Drosophila". Trends in Genetics. 19 (6): 310–1. doi:10.1016/S0168-9525(03)00050-7. PMID 12801722.
- West-Eberhard MJ (May 2005). "Developmental plasticity and the origin of species differences". Proceedings of the National Academy of Sciences of the United States of America. 102 Suppl 1 (suppl 1): 6543–9. Bibcode:2005PNAS..102.6543W. doi:10.1073/pnas.0501844102. PMC 1131862. PMID 15851679.
- Abram PK, Boivin G, Moiroux J, Brodeur J (November 2017). "Behavioural effects of temperature on ectothermic animals: unifying thermal physiology and behavioural plasticity". Biological Reviews of the Cambridge Philosophical Society. 92 (4): 1859–1876. doi:10.1111/brv.12312. PMID 28980433. S2CID 9099834.
- Gibert P, Huey RB, Gilchrist GW (January 2001). "Locomotor performance of Drosophila melanogaster: interactions among developmental and adult temperatures, age, and geography". Evolution; International Journal of Organic Evolution. 55 (1): 205–9. doi:10.1111/j.0014-3820.2001.tb01286.x. PMID 11263741. S2CID 2991855.
- Zamudio KR, Huey RB, Crill WD (1995). "Bigger isn't always better: body size, developmental and parental temperature and male territorial success in Drosophila melanogaster". Animal Behaviour. 49 (3): 671–677. doi:10.1016/0003-3472(95)80200-2. ISSN 0003-3472. S2CID 9124942.
- Harrison JF, Waters JS, Biddulph TA, Kovacevic A, Klok CJ, Socha JJ (April 2018). "Developmental plasticity and stability in the tracheal networks supplying Drosophila flight muscle in response to rearing oxygen level". Journal of Insect Physiology. The limits of respiratory function: external and internal constraints on insect gas exchange. 106 (Pt 3): 189–198. doi:10.1016/j.jinsphys.2017.09.006. PMID 28927826.
- Sheeba V, Chandrashekaran MK, Joshi A, Sharma VK (January 2002). "Developmental plasticity of the locomotor activity rhythm of Drosophila melanogaster". Journal of Insect Physiology. 48 (1): 25–32. doi:10.1016/S0022-1910(01)00139-1. PMID 12770129.
- Crill WD, Huey RB, Gilchrist GW (June 1996). "Within- and Between-Generation Effects of Temperature on the Morphology and Physiology of Drosophila Melanogaster". Evolution; International Journal of Organic Evolution. 50 (3): 1205–1218. doi:10.2307/2410661. JSTOR 2410661. PMID 28565273.
- David JR, Araripe LO, Chakir M, Legout H, Lemos B, Pétavy G, et al. (July 2005). "Male sterility at extreme temperatures: a significant but neglected phenomenon for understanding Drosophila climatic adaptations". Journal of Evolutionary Biology. 18 (4): 838–46. doi:10.1111/j.1420-9101.2005.00914.x. PMID 16033555. S2CID 23847613.
- French V, Feast M, Partridge L (November 1998). "Body size and cell size in Drosophila: the developmental response to temperature". Journal of Insect Physiology. 44 (11): 1081–1089. doi:10.1016/S0022-1910(98)00061-4. PMID 12770407.
- Frazier MR, Harrison JF, Kirkton SD, Roberts SP (July 2008). "Cold rearing improves cold-flight performance in Drosophila via changes in wing morphology". The Journal of Experimental Biology. 211 (Pt 13): 2116–22. doi:10.1242/jeb.019422. PMID 18552301.
- Slotsbo S, Schou MF, Kristensen TN, Loeschcke V, Sørensen JG (September 2016). "Reversibility of developmental heat and cold plasticity is asymmetric and has long-lasting consequences for adult thermal tolerance". The Journal of Experimental Biology. 219 (Pt 17): 2726–32. doi:10.1242/jeb.143750. PMID 27353229.
- Gilchrist GW, Huey RB (January 2001). "Parental and developmental temperature effects on the thermal dependence of fitness in Drosophila melanogaster". Evolution; International Journal of Organic Evolution. 55 (1): 209–14. doi:10.1111/j.0014-3820.2001.tb01287.x. PMID 11263742. S2CID 1329035.
- Austin CJ, Moehring AJ (May 2013). "Optimal temperature range of a plastic species, Drosophila simulans". The Journal of Animal Ecology. 82 (3): 663–72. doi:10.1111/1365-2656.12041. PMID 23360477.
- Hodin J, Riddiford LM (October 2000). "Different mechanisms underlie phenotypic plasticity and interspecific variation for a reproductive character in drosophilids (Insecta: Diptera)". Evolution; International Journal of Organic Evolution. 54 (5): 1638–53. doi:10.1111/j.0014-3820.2000.tb00708.x. PMID 11108591. S2CID 6875815.
- Klepsatel P, Girish TN, Dircksen H, Gáliková M (May 2019). "Drosophila is maximised by optimal developmental temperature". The Journal of Experimental Biology. 222 (Pt 10): jeb202184. doi:10.1242/jeb.202184. PMID 31064855.
- Schou MF, Kristensen TN, Pedersen A, Karlsson BG, Loeschcke V, Malmendal A (February 2017). "Metabolic and functional characterization of effects of developmental temperature in Drosophila melanogaster". American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 312 (2): R211–R222. doi:10.1152/ajpregu.00268.2016. PMC 5336569. PMID 27927623.
- Cohet Y, David J (January 1978). "Control of the adult reproductive potential by preimaginal thermal conditions: A study in Drosophila melanogaster". Oecologia. 36 (3): 295–306. Bibcode:1978Oecol..36..295C. doi:10.1007/BF00348055. PMID 28309916. S2CID 12465060.
- Rideout EJ, Narsaiya MS, Grewal SS (December 2015). "The Sex Determination Gene transformer Regulates Male-Female Differences in Drosophila Body Size". PLOS Genetics. 11 (12): e1005683. doi:10.1371/journal.pgen.1005683. PMC 4692505. PMID 26710087.
- Gilbert SF (2000). Developmental Biology (6th ed.). Sunderland (MA): Sinauer Associates; 2000. ISBN 9780878932436.
- Lemaitre B, Hoffmann J (2007). "The host defense of Drosophila melanogaster" (PDF). Annual Review of Immunology. 25: 697–743. doi:10.1146/annurev.immunol.25.022106.141615. PMID 17201680.
- Troha K, Im JH, Revah J, Lazzaro BP, Buchon N (February 2018). "Comparative transcriptomics reveals CrebA as a novel regulator of infection tolerance in D. melanogaster". PLOS Pathogens. 14 (2): e1006847. doi:10.1371/journal.ppat.1006847. PMC 5812652. PMID 29394281.
- De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B (June 2002). "The Toll and Imd pathways are the major regulators of the immune response in Drosophila". The EMBO Journal. 21 (11): 2568–79. doi:10.1093/emboj/21.11.2568. PMC 126042. PMID 12032070.
- Paredes JC, Welchman DP, Poidevin M, Lemaitre B (November 2011). "Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection" (PDF). Immunity. 35 (5): 770–9. doi:10.1016/j.immuni.2011.09.018. PMID 22118526.
- Troha K, Buchon N (September 2019). "Methods for the study of innate immunity in Drosophila melanogaster". Wiley Interdisciplinary Reviews. Developmental Biology. 8 (5): e344. doi:10.1002/wdev.344. PMID 30993906. S2CID 119527642.
- Gilbert R, Torres M, Clemens R, Hateley S, Hosamani R, Wade W, Bhattacharya S (February 2020). "Drosophila melanogaster infection model". NPJ Microgravity. 6 (1): 4. doi:10.1038/s41526-019-0091-2. PMC 7000411. PMID 32047838.
- Valanne S, Wang JH, Rämet M (January 2011). "The Drosophila Toll signaling pathway". Journal of Immunology. 186 (2): 649–56. doi:10.4049/jimmunol.1002302. PMID 21209287.
- Dudzic JP, Hanson MA, Iatsenko I, Kondo S, Lemaitre B (April 2019). "More Than Black or White: Melanization and Toll Share Regulatory Serine Proteases in Drosophila". Cell Reports. 27 (4): 1050–1061.e3. doi:10.1016/j.celrep.2019.03.101. PMID 31018123.
- Hanson MA, Hamilton PT, Perlman SJ (October 2016). "Immune genes and divergent antimicrobial peptides in flies of the subgenus Drosophila". BMC Evolutionary Biology. 16 (1): 228. doi:10.1186/s12862-016-0805-y. PMC 5078906. PMID 27776480.
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA (September 1996). "The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults" (PDF). Cell. 86 (6): 973–83. doi:10.1016/s0092-8674(00)80172-5. PMID 8808632. S2CID 10736743.
- "The Nobel Prize in Physiology or Medicine 2011". NobelPrize.org. Retrieved 2020-09-05.
- Issa N, Guillaumot N, Lauret E, Matt N, Schaeffer-Reiss C, Van Dorsselaer A, et al. (February 2018). "The Circulating Protease Persephone Is an Immune Sensor for Microbial Proteolytic Activities Upstream of the Drosophila Toll Pathway". Molecular Cell. 69 (4): 539–550.e6. doi:10.1016/j.molcel.2018.01.029. PMC 5823974. PMID 29452635.
- Hetru, Charles; Hoffmann, Jules A. (2009). "NF-kappaB in the immune response of Drosophila". Cold Spring Harbor Perspectives in Biology. 1 (6): a000232. doi:10.1101/cshperspect.a000232. ISSN 1943-0264. PMC 2882123. PMID 20457557.
- Dudzic, Jan Paul; Hanson, Mark Austin; Iatsenko, Igor; Kondo, Shu; Lemaitre, Bruno (2019-04-23). "More Than Black or White: Melanization and Toll Share Regulatory Serine Proteases in Drosophila". Cell Reports. 27 (4): 1050–1061.e3. doi:10.1016/j.celrep.2019.03.101. ISSN 2211-1247. PMID 31018123. S2CID 131775641.
- Belmonte, Rebecca L.; Corbally, Mary-Kate; Duneau, David F.; Regan, Jennifer C. (2020-01-31). "Sexual Dimorphisms in Innate Immunity and Responses to Infection in Drosophila melanogaster". Frontiers in Immunology. Frontiers Media. 10: 3075. doi:10.3389/fimmu.2019.03075. ISSN 1664-3224. PMC 7006818. PMID 32076419.
- Cerenius, Lage; Söderhäll, Kenneth (2021). "Immune properties of invertebrate phenoloxidases". Developmental & Comparative Immunology. Elsevier. 122: 104098. doi:10.1016/j.dci.2021.104098. ISSN 0145-305X. PMID 33857469.
- Clemmons AW, Lindsay SA, Wasserman SA (April 2015). "An effector Peptide family required for Drosophila toll-mediated immunity". PLOS Pathogens. 11 (4): e1004876. doi:10.1371/journal.ppat.1004876. PMC 4411088. PMID 25915418.
- Lindsay SA, Lin SJ, Wasserman SA (2018). "Short-Form Bomanins Mediate Humoral Immunity in Drosophila". Journal of Innate Immunity. 10 (4): 306–314. doi:10.1159/000489831. PMC 6158068. PMID 29920489.
- Yamamoto-Hino M, Goto S (May 2016). "Spätzle-Processing Enzyme-independent Activation of the Toll Pathway in Drosophila Innate Immunity". Cell Structure and Function. 41 (1): 55–60. doi:10.1247/csf.16002. PMID 26843333.
- Troha K, Nagy P, Pivovar A, Lazzaro BP, Hartley PS, Buchon N (October 2019). "Nephrocytes Remove Microbiota-Derived Peptidoglycan from Systemic Circulation to Maintain Immune Homeostasis". Immunity. 51 (4): 625–637.e3. doi:10.1016/j.immuni.2019.08.020. PMID 31564469.
- Hanson MA, Dostálová A, Ceroni C, Poidevin M, Kondo S, Lemaitre B (February 2019). "Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach". eLife. 8. doi:10.7554/eLife.44341. PMC 6398976. PMID 30803481.
- Zhai Z, Boquete JP, Lemaitre B (June 2017). "A genetic framework controlling the differentiation of intestinal stem cells during regeneration in Drosophila". PLOS Genetics. 13 (6): e1006854. doi:10.1371/journal.pgen.1006854. PMC 5510897. PMID 28662029.
- Nandy A, Lin L, Velentzas PD, Wu LP, Baehrecke EH, Silverman N (November 2018). "The NF-κB Factor Relish Regulates Atg1 Expression and Controls Autophagy". Cell Reports. 25 (8): 2110–2120.e3. doi:10.1016/j.celrep.2018.10.076. PMC 6329390. PMID 30463009.
- Hanson MA, Lemaitre B (February 2020). "New insights on Drosophila antimicrobial peptide function in host defense and beyond". Current Opinion in Immunology. 62: 22–30. doi:10.1016/j.coi.2019.11.008. PMID 31835066.
- Kounatidis I, Chtarbanova S (2018). "Drosophila Perspective". Frontiers in Immunology. 9: 1362. doi:10.3389/fimmu.2018.01362. PMC 6004738. PMID 29942319.
- Cao Y, Chtarbanova S, Petersen AJ, Ganetzky B (May 2013). "Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain". Proceedings of the National Academy of Sciences of the United States of America. 110 (19): E1752-60. Bibcode:2013PNAS..110E1752C. doi:10.1073/pnas.1306220110. PMC 3651420. PMID 23613578.
- Petersen AJ, Katzenberger RJ, Wassarman DA (May 2013). "The innate immune response transcription factor relish is necessary for neurodegeneration in a Drosophila model of ataxia-telangiectasia". Genetics. 194 (1): 133–42. doi:10.1534/genetics.113.150854. PMC 3632461. PMID 23502677.
- Kounatidis I, Chtarbanova S, Cao Y, Hayne M, Jayanth D, Ganetzky B, Ligoxygakis P (April 2017). "NF-κB Immunity in the Brain Determines Fly Lifespan in Healthy Aging and Age-Related Neurodegeneration". Cell Reports. 19 (4): 836–848. doi:10.1016/j.celrep.2017.04.007. PMC 5413584. PMID 28445733.
- Parvy JP, Yu Y, Dostalova A, Kondo S, Kurjan A, Bulet P, et al. (July 2019). "Drosophila". eLife. 8. doi:10.7554/eLife.45061. PMC 6667213. PMID 31358113.
- Araki M, Kurihara M, Kinoshita S, Awane R, Sato T, Ohkawa Y, Inoue YH (June 2019). "Drosophila mxc mutants". Disease Models & Mechanisms. 12 (6). doi:10.1242/dmm.037721. PMC 6602314. PMID 31160313.
- Barajas-Azpeleta R, Wu J, Gill J, Welte R, Seidel C, McKinney S, et al. (October 2018). "Antimicrobial peptides modulate long-term memory". PLOS Genetics. 14 (10): e1007440. doi:10.1371/journal.pgen.1007440. PMC 6224176. PMID 30312294.
- Dostálová A, Rommelaere S, Poidevin M, Lemaitre B (September 2017). "Thioester-containing proteins regulate the Toll pathway and play a role in Drosophila defence against microbial pathogens and parasitoid wasps". BMC Biology. 15 (1): 79. doi:10.1186/s12915-017-0408-0. PMC 5584532. PMID 28874153.
- Srinivasan N, Gordon O, Ahrens S, Franz A, Deddouche S, Chakravarty P, et al. (November 2016). "Drosophila melanogaster". eLife. 5. doi:10.7554/eLife.19662. PMC 5138034. PMID 27871362.
- Goto A, Yano T, Terashima J, Iwashita S, Oshima Y, Kurata S (May 2010). "Cooperative regulation of the induction of the novel antibacterial Listericin by peptidoglycan recognition protein LE and the JAK-STAT pathway". The Journal of Biological Chemistry. 285 (21): 15731–8. doi:10.1074/jbc.M109.082115. PMC 2871439. PMID 20348097.
- Wang L, Kounatidis I, Ligoxygakis P (January 2014). "Drosophila as a model to study the role of blood cells in inflammation, innate immunity and cancer". Frontiers in Cellular and Infection Microbiology. 3: 113. doi:10.3389/fcimb.2013.00113. PMC 3885817. PMID 24409421.
- Neyen C, Bretscher AJ, Binggeli O, Lemaitre B (June 2014). "Methods to study Drosophila immunity" (PDF). Methods. 68 (1): 116–28. doi:10.1016/j.ymeth.2014.02.023. PMID 24631888.
- Hashimoto Y, Tabuchi Y, Sakurai K, Kutsuna M, Kurokawa K, Awasaki T, et al. (December 2009). "Identification of lipoteichoic acid as a ligand for draper in the phagocytosis of Staphylococcus aureus by Drosophila hemocytes". Journal of Immunology. 183 (11): 7451–60. doi:10.4049/jimmunol.0901032. PMID 19890048.
- Holz A, Bossinger B, Strasser T, Janning W, Klapper R (October 2003). "The two origins of hemocytes in Drosophila". Development. 130 (20): 4955–62. doi:10.1242/dev.00702. PMID 12930778.
- Sanchez Bosch P, Makhijani K, Herboso L, Gold KS, Baginsky R, Woodcock KJ, et al. (December 2019). "Adult Drosophila Lack Hematopoiesis but Rely on a Blood Cell Reservoir at the Respiratory Epithelia to Relay Infection Signals to Surrounding Tissues". Developmental Cell. 51 (6): 787–803.e5. doi:10.1016/j.devcel.2019.10.017. PMC 7263735. PMID 31735669.
- Parvy JP, Yu Y, Dostalova A, Kondo S, Kurjan A, Bulet P, et al. (July 2019). "Drosophila". eLife. 8: e45061. doi:10.7554/eLife.45061. PMC 6667213. PMID 31358113.
- Sturtevant AH (1929). "The claret mutant type of Drosophila simulans: a study of chromosome elimination and cell-lineage". Zeitschrift für Wissenschaftliche Zoologie. 135: 323–356.
- Nissani M (May 1975). "A new behavioral bioassay for an analysis of sexual attraction and pheromones in insects". The Journal of Experimental Zoology. 192 (2): 271–5. doi:10.1002/jez.1401920217. PMID 805823.
- Khan FA (2011). Biotechnology Fundamentals. CRC Press. p. 213. ISBN 978-1-4398-2009-4.
- "The 2017 Nobel Prize in Physiology or Medicine jointly to Jeffrey C. Hall, Michael Rosbash and Michael W. Young for their discoveries of molecular mechanisms controlling the circadian rhythm". Nobelprize.org. 2 October 2017. Retrieved 5 October 2017.
- Lehnert BP, Baker AE, Gaudry Q, Chiang AS, Wilson RI (January 2013). "Distinct roles of TRP channels in auditory transduction and amplification in Drosophila". Neuron. 77 (1): 115–28. doi:10.1016/j.neuron.2012.11.030. PMC 3811118. PMID 23312520.
- Zhang W, Yan Z, Jan LY, Jan YN (August 2013). "Sound response mediated by the TRP channels NOMPC, NANCHUNG, and INACTIVE in chordotonal organs of Drosophila larvae". Proceedings of the National Academy of Sciences of the United States of America. 110 (33): 13612–7. Bibcode:2013PNAS..11013612Z. doi:10.1073/pnas.1312477110. PMC 3746866. PMID 23898199.
- "Homosexuality Turned On and Off in Fruit Flies"
- Dasgupta S, Sheehan TC, Stevens CF, Navlakha S (December 2018). "A neural data structure for novelty detection". Proceedings of the National Academy of Sciences of the United States of America. 115 (51): 13093–13098. Bibcode:2018PNAS..11513093D. doi:10.1073/pnas.1814448115. PMC 6304992. PMID 30509984.
- Zwarts L, Versteven M, Callaerts P (2012-01-01). "Genetics and neurobiology of aggression in Drosophila". Fly. 6 (1): 35–48. doi:10.4161/fly.19249. PMC 3365836. PMID 22513455.
- Davis SM, Thomas AL, Liu L, Campbell IM, Dierick HA (January 2018). "Drosophila Using a Screen for Wing Damage". Genetics. 208 (1): 273–282. doi:10.1534/genetics.117.300292. PMC 5753862. PMID 29109180.
- Versteven M, Vanden Broeck L, Geurten B, Zwarts L, Decraecker L, Beelen M, et al. (February 2017). "Drosophila aggression". Proceedings of the National Academy of Sciences of the United States of America. 114 (8): 1958–1963. doi:10.1073/pnas.1605946114. PMC 5338383. PMID 28115690.
- Sengupta S, Smith DP (2014). Mucignat-Caretta C (ed.). How Drosophila Detect Volatile Pheromones: Signaling, Circuits, and Behavior. Neurobiology of Chemical Communication. Frontiers in Neuroscience. CRC Press/Taylor & Francis. ISBN 9781466553415. PMID 24830032. Retrieved 2019-05-30.
- Laturney M, Billeter JC (August 2016). "Drosophila melanogaster females restore their attractiveness after mating by removing male anti-aphrodisiac pheromones". Nature Communications. 7 (1): 12322. Bibcode:2016NatCo...712322L. doi:10.1038/ncomms12322. PMC 4976142. PMID 27484362.
- Wang L, Han X, Mehren J, Hiroi M, Billeter JC, Miyamoto T, et al. (June 2011). "Hierarchical chemosensory regulation of male-male social interactions in Drosophila". Nature Neuroscience. 14 (6): 757–62. doi:10.1038/nn.2800. PMC 3102769. PMID 21516101.
- Lim RS, Eyjólfsdóttir E, Shin E, Perona P, Anderson DJ (2014-08-27). "How food controls aggression in Drosophila". PLOS ONE. 9 (8): e105626. Bibcode:2014PLoSO...9j5626L. doi:10.1371/journal.pone.0105626. PMC 4146546. PMID 25162609.
- Erion R, DiAngelo JR, Crocker A, Sehgal A (September 2012). "Interaction between sleep and metabolism in Drosophila with altered octopamine signaling". The Journal of Biological Chemistry. 287 (39): 32406–14. doi:10.1074/jbc.M112.360875. PMC 3463357. PMID 22829591.
- Kayser MS, Mainwaring B, Yue Z, Sehgal A (July 2015). Griffith LC (ed.). "Sleep deprivation suppresses aggression in Drosophila". eLife. 4: e07643. doi:10.7554/eLife.07643. PMC 4515473. PMID 26216041.
- Hughes TT, Allen AL, Bardin JE, Christian MN, Daimon K, Dozier KD, et al. (February 2012). "Drosophila as a genetic model for studying pathogenic human viruses". Virology. 423 (1): 1–5. doi:10.1016/j.virol.2011.11.016. PMC 3253880. PMID 22177780.
- Hardie RC, Raghu P (September 2001). "Visual transduction in Drosophila". Nature. 413 (6852): 186–93. Bibcode:2001Natur.413..186H. doi:10.1038/35093002. PMID 11557987. S2CID 4415605.
- Sharkey, Camilla R.; Blanco, Jorge; Leibowitz, Maya M.; Pinto-Benito, Daniel; Wardill, Trevor J. (December 2020). "The spectral sensitivity of Drosophila photoreceptors". Scientific Reports. 10 (1): 18242. doi:10.1038/s41598-020-74742-1. PMID 33106518.
 Material was copied and adapted from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied and adapted from this source, which is available under a Creative Commons Attribution 4.0 International License. - Feuda, Roberto; Goulty, Matthew; Zadra, Nicola; Gasparetti, Tiziana; Rosato, Ezio; Pisani, Davide; Rizzoli, Annapaola; Segata, Nicola; Ometto, Lino; Stabelli, Omar Rota (3 August 2021). "Phylogenomics of Opsin Genes in Diptera Reveals Lineage-Specific Events and Contrasting Evolutionary Dynamics in Anopheles and Drosophila". Genome Biology and Evolution. 13 (8): evab170. doi:10.1093/gbe/evab170. PMID 34270718.
- Stark, William S.; Walker, John A.; Harris, William A. (1 April 1976). "Genetic dissection of the photoreceptor system in the compound eye of Drosophila melanogaster". The Journal of Physiology. 256 (2): 415–439. doi:10.1113/jphysiol.1976.sp011331. PMID 16992509.
- Ostroy, Sanford E.; Wilson, Meegan; Pak, William L. (August 1974). "Drosophila rhodopsin: Photochemistry, extraction and differences in the norp AP12 phototransduction mutant". Biochemical and Biophysical Research Communications. 59 (3): 960–966. doi:10.1016/s0006-291x(74)80073-2. PMID 4213042.
- Ostroy, S E (1 November 1978). "Characteristics of Drosophila rhodopsin in wild-type and norpA vision transduction mutants". Journal of General Physiology. 72 (5): 717–732. doi:10.1085/jgp.72.5.717. PMID 105082.
- Salcedo, Ernesto; Huber, Armin; Henrich, Stefan; Chadwell, Linda V.; Chou, Wen-Hai; Paulsen, Reinhard; Britt, Steven G. (15 December 1999). "Blue- and Green-Absorbing Visual Pigments of Drosophila: Ectopic Expression and Physiological Characterization of the R8 Photoreceptor Cell-Specific Rh5 and Rh6 Rhodopsins". The Journal of Neuroscience. 19 (24): 10716–10726. doi:10.1523/jneurosci.19-24-10716.1999. PMID 10594055.
- Kirschfeld, K.; Franceschini, N.; Minke, B. (September 1977). "Evidence for a sensitising pigment in fly photoreceptors". Nature. 269 (5627): 386–390. doi:10.1038/269386a0.
- Minke, B; Kirschfeld, K (May 1979). "The contribution of a sensitizing pigment to the photosensitivity spectra of fly rhodopsin and metarhodopsin". The Journal of general physiology. 73 (5): 517–40. doi:10.1085/jgp.73.5.517. PMID 458418.
- Kirschfeld, K.; Feiler, R.; Franceschini, N. (1978). "A photostable pigment within the rhabdomere of fly photoreceptors no. 7". Journal of Comparative Physiology ? A. 125 (3): 275–284. doi:10.1007/BF00656606.
- Kirschfeld, K.; Franceschini, N. (1977). "Photostable pigments within the membrane of photoreceptors and their possible role". Biophysics of Structure and Mechanism. 3 (2): 191–194. doi:10.1007/BF00535818. PMID 890056.
- Zuker, CS; Montell, C; Jones, K; Laverty, T; Rubin, GM (1 May 1987). "A rhodopsin gene expressed in photoreceptor cell R7 of the Drosophila eye: homologies with other signal-transducing molecules". The Journal of Neuroscience. 7 (5): 1550–1557. doi:10.1523/jneurosci.07-05-01550.1987. PMID 2437266.
- Chou, Wen-Hai; Hall, Kristin J; Wilson, D.Bianca; Wideman, Christi L; Townson, Steven M; Chadwell, Linda V; Britt, Steven G (December 1996). "Identification of a Novel Drosophila Opsin Reveals Specific Patterning of the R7 and R8 Photoreceptor Cells". Neuron. 17 (6): 1101–1115. doi:10.1016/s0896-6273(00)80243-3. PMID 8982159.
- Feiler, R; Bjornson, R; Kirschfeld, K; Mismer, D; Rubin, Gm; Smith, Dp; Socolich, M; Zuker, Cs (1 October 1992). "Ectopic expression of ultraviolet-rhodopsins in the blue photoreceptor cells of Drosophila: visual physiology and photochemistry of transgenic animals". The Journal of Neuroscience. 12 (10): 3862–3868. doi:10.1523/jneurosci.12-10-03862.1992.
- Montell, C; Jones, K; Zuker, C; Rubin, G (1 May 1987). "A second opsin gene expressed in the ultraviolet-sensitive R7 photoreceptor cells of Drosophila melanogaster". The Journal of Neuroscience. 7 (5): 1558–1566. doi:10.1523/JNEUROSCI.07-05-01558.1987. PMID 2952772.
- Huber, Armin; Schulz, Simone; Bentrop, Joachim; Groell, Christine; Wolfrum, Uwe; Paulsen, Reinhard (7 April 1997). "Molecular cloning of Drosophila Rh6 rhodopsin: the visual pigment of a subset of R8 photoreceptor cells". FEBS Letters. 406 (1–2): 6–10. doi:10.1016/s0014-5793(97)00210-x. PMID 9109375.
- Pollock, John A.; Benzer, Seymour (June 1988). "Transcript localization of four opsin genes in the three visual organs of Drosophila; RH2 is ocellus specific". Nature. 333 (6175): 779–782. doi:10.1038/333779a0. PMID 2968518.
- Feiler, Reinhard; Harris, William A.; Kirschfeld, Kuno; Wehrhahn, Christian; Zuker, Charles S. (June 1988). "Targeted misexpression of a Drosophila opsin gene leads to altered visual function". Nature. 333 (6175): 737–741. doi:10.1038/333737a0.
- Sakai, Kazumi; Tsutsui, Kei; Yamashita, Takahiro; Iwabe, Naoyuki; Takahashi, Keisuke; Wada, Akimori; Shichida, Yoshinori (December 2017). "Drosophila melanogaster rhodopsin Rh7 is a UV-to-visible light sensor with an extraordinarily broad absorption spectrum". Scientific Reports. 7 (1): 7349. doi:10.1038/s41598-017-07461-9. PMID 28779161.
- Ni, Jinfei D.; Baik, Lisa S.; Holmes, Todd C.; Montell, Craig (May 2017). "A rhodopsin in the brain functions in circadian photoentrainment in Drosophila". Nature. 545 (7654): 340–344. doi:10.1038/nature22325. PMID 28489826.
- Vogt, Klaus (1 February 1984). "The Chromophore of the Visual Pigment in Some Insect Orders". Zeitschrift für Naturforschung C. 39 (1–2): 196–197. doi:10.1515/znc-1984-1-236.
- Vogt, K.; Kirschfeld, K. (April 1984). "Chemical identity of the chromophores of fly visual pigment". Naturwissenschaften. 71 (4): 211–213. doi:10.1007/BF00490436.
- Gühmann M, Porter ML, Bok MJ (August 2022). "The Gluopsins: Opsins without the Retinal Binding Lysine". Cells. 11 (15): 2441. doi:10.3390/cells11152441. PMC 9368030. PMID 35954284.
- Katana R, Guan C, Zanini D, Larsen ME, Giraldo D, Geurten BR, et al. (September 2019). "Chromophore-Independent Roles of Opsin Apoproteins in Drosophila Mechanoreceptors". Current Biology. 29 (17): 2961–2969.e4. doi:10.1016/j.cub.2019.07.036. PMID 31447373. S2CID 201420079.
- Leung NY, Thakur DP, Gurav AS, Kim SH, Di Pizio A, Niv MY, Montell C (April 2020). "Functions of Opsins in Drosophila Taste". Current Biology. 30 (8): 1367–1379.e6. doi:10.1016/j.cub.2020.01.068. PMC 7252503. PMID 32243853.
- Kumbalasiri T, Rollag MD, Isoldi MC, Castrucci AM, Provencio I (March 2007). "Melanopsin triggers the release of internal calcium stores in response to light". Photochemistry and Photobiology. 83 (2): 273–279. doi:10.1562/2006-07-11-RA-964. PMID 16961436.
- Raghu P, Colley NJ, Webel R, James T, Hasan G, Danin M, et al. (May 2000). "Normal phototransduction in Drosophila photoreceptors lacking an InsP(3) receptor gene". Molecular and Cellular Neurosciences. 15 (5): 429–45. doi:10.1006/mcne.2000.0846. PMID 10833300. S2CID 23861204.
- Wang T, Xu H, Oberwinkler J, Gu Y, Hardie RC, Montell C (February 2005). "Light activation, adaptation, and cell survival functions of the Na+/Ca2+ exchanger CalX". Neuron. 45 (3): 367–78. doi:10.1016/j.neuron.2004.12.046. PMID 15694324.
- Rein K, Zöckler M, Mader MT, Grübel C, Heisenberg M (February 2002). "The Drosophila standard brain". Current Biology. 12 (3): 227–31. doi:10.1016/S0960-9822(02)00656-5. PMID 11839276. S2CID 15785406.
- Zhu, Huanhu; Han, Min (2014-11-23). "Exploring Developmental and Physiological Functions of Fatty Acid and Lipid Variants Through Worm and Fly Genetics". Annual Review of Genetics. Annual Reviews. 48 (1): 119–148. doi:10.1146/annurev-genet-041814-095928. ISSN 0066-4197. PMID 25195508.
- Dawkins R, Dawkins M (1976). "Hierarchical organization and postural facilitation: rules for grooming in flies". Animal Behaviour. 24 (4): 739–755. doi:10.1016/S0003-3472(76)80003-6. S2CID 53186674.
- Davis WJ (1979). "Behavioural hierarchies". Trends in Neurosciences. 2 (2): 5–7. doi:10.1016/0166-2236(79)90003-1. S2CID 53180462.
- Seeds AM, Ravbar P, Chung P, Hampel S, Midgley FM, Mensh BD, Simpson JH (August 2014). "A suppression hierarchy among competing motor programs drives sequential grooming in Drosophila". eLife. 3: e02951. doi:10.7554/eLife.02951. PMC 4136539. PMID 25139955.
- Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, Bethge M (September 2018). "DeepLabCut: markerless pose estimation of user-defined body parts with deep learning". Nature Neuroscience. 21 (9): 1281–1289. doi:10.1038/s41593-018-0209-y. PMID 30127430. S2CID 4748395.
- Strauss R, Heisenberg M (August 1990). "Coordination of legs during straight walking and turning in Drosophila melanogaster". Journal of Comparative Physiology A: Sensory, Neural, and Behavioral Physiology. 167 (3): 403–12. doi:10.1007/BF00192575. PMID 2121965. S2CID 12965869.
- DeAngelis BD, Zavatone-Veth JA, Clark DA (June 2019). "Drosophila". eLife. 8. doi:10.7554/eLife.46409. PMC 6598772. PMID 31250807.
- Wosnitza A, Bockemühl T, Dübbert M, Scholz H, Büschges A (February 2013). "Inter-leg coordination in the control of walking speed in Drosophila". The Journal of Experimental Biology. 216 (Pt 3): 480–91. doi:10.1242/jeb.078139. PMID 23038731.
- Mendes CS, Bartos I, Akay T, Márka S, Mann RS (January 2013). "Quantification of gait parameters in freely walking wild type and sensory deprived Drosophila melanogaster". eLife. 2: e00231. doi:10.7554/eLife.00231. PMC 3545443. PMID 23326642.
- Szczecinski NS, Bockemühl T, Chockley AS, Büschges A (November 2018). "Drosophila". The Journal of Experimental Biology. 221 (Pt 22): jeb189142. doi:10.1242/jeb.189142. PMID 30274987.
- Hooper SL (May 2012). "Body size and the neural control of movement". Current Biology. 22 (9): R318-22. doi:10.1016/j.cub.2012.02.048. PMID 22575473.
- Fry SN, Sayaman R, Dickinson MH (April 2003). "The aerodynamics of free-flight maneuvers in Drosophila" (PDF). Science. 300 (5618): 495–8. Bibcode:2003Sci...300..495F. doi:10.1126/science.1081944. PMID 12702878. S2CID 40952385. Archived from the original (PDF) on 2015-09-24.
- Hesselberg T, Lehmann FO (December 2007). "Turning behaviour depends on frictional damping in the fruit fly Drosophila". The Journal of Experimental Biology. 210 (Pt 24): 4319–34. doi:10.1242/jeb.010389. PMID 18055621.
- "Non pest species". Plant Health Australia. Retrieved September 19, 2017.
- McEvey S (February 5, 2014). "Fruit Flies: A Case Of Mistaken Identity". Australian Museum. Retrieved September 19, 2017.
Further reading
- Kohler RE (1994). Lords of the Fly: Drosophila genetics and the experimental life. Chicago: University of Chicago Press. ISBN 978-0-226-45063-6.
- Gilbert SF (2000). Developmental Biology (6th ed.). Sunderland (MA): Sinauer Associates; 2000. ISBN 9780878932436.
- Perrimon N, Bonini NM, Dhillon P (March 2016). "Fruit flies on the front line: the translational impact of Drosophila". Disease Models & Mechanisms. 9 (3): 229–31. doi:10.1242/dmm.024810. PMC 4833334. PMID 26935101.
- Henderson M (April 8, 2010). "Row over fruit fly Drosophila melanogaster name bugs scientists". The Times. The Australian. Retrieved September 19, 2017.
External links
- "A quick and simple introduction to Drosophila melanogaster". Drosophila Virtual Library.
- "Drosophila Genomics Resource Center" – collects, maintains and distributes Drosophila DNA clones and cell lines.
- "Bloomington Drosophila Stock Center" – collects, maintains and distributes Drosophila melanogaster strains for research
- "FlyBase—A Database of Drosophila Genes & Genomes".
- "NCBI Map Viewer – Drosophila melanogaster".
- "Drosophila Virtual Library".
- "The Berkeley Drosophila Genome Project".
- "FlyMove". – video resources for Drosophila development
- "Drosophila Nomenclature—naming of genes". Archived from the original on 8 October 2011.
- View the Fruitfly genome on Ensembl
- View the dm6 genome assembly in the UCSC Genome Browser.
- Manchester Fly Facility – for the public from the University of Manchester
- The droso4schools website with school-relevant resources about Drosophila
- Part 1 of the "Small fly: BIG impact" educational videos explaining the history and importance of the model organism Drosophila.
- Part 2 of the "Small fly: BIG impact" educational videos explaining how research is carried out in Drosophila.
- "Inside the Fly Lab"—broadcast by WGBH and PBS, in the program series Curious, January 2008.
- "How a Fly Detects Poison"—WhyFiles.org article describes how the fruit fly tastes a larva-killing chemical in food.