Drosophila
Drosophila (/drəˈsɒfɪlə, drɒ-, droʊ-/[1][2]) is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species to linger around overripe or rotting fruit. They should not be confused with the Tephritidae, a related family, which are also called fruit flies (sometimes referred to as "true fruit flies"); tephritids feed primarily on unripe or ripe fruit, with many species being regarded as destructive agricultural pests, especially the Mediterranean fruit fly.
| Drosophila | |
|---|---|
 | |
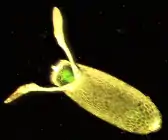
| Drosophila pseudoobscura | |
| Scientific classification | |
| Kingdom: | Animalia |
| Phylum: | Arthropoda |
| Class: | Insecta |
| Order: | Diptera |
| Family: | Drosophilidae |
| Subfamily: | Drosophilinae |
| Genus: | Drosophila Fallén, 1823 |
| Type species | |
| Musca funebris Fabricius, 1787 | |
| Subgenera | |
| |
| Synonyms | |
|
Oinopota Kirby & Spence, 1815 | |
One species of Drosophila in particular, D. melanogaster, has been heavily used in research in genetics and is a common model organism in developmental biology. The terms "fruit fly" and "Drosophila" are often used synonymously with D. melanogaster in modern biological literature. The entire genus, however, contains more than 1,500 species[3] and is very diverse in appearance, behavior, and breeding habitat.
Etymology
The term "Drosophila", meaning "dew-loving", is a modern scientific Latin adaptation from Greek words δρόσος, drósos, "dew", and φίλος, phílos, "loving" with the Latin feminine suffix -a.
Morphology
Drosophila species are small flies, typically pale yellow to reddish brown to black, with red eyes. When the eyes (essentially a film of lenses) are removed, the brain is revealed. Drosophila brain structure and function develop and age significantly from larval to adult stage. Developing brain structures make these flies a prime candidate for neuro-genetic research.[4] Many species, including the noted Hawaiian picture-wings, have distinct black patterns on the wings. The plumose (feathery) arista, bristling of the head and thorax, and wing venation are characters used to diagnose the family. Most are small, about 2–4 millimetres (0.079–0.157 in) long, but some, especially many of the Hawaiian species, are larger than a house fly.
Evolution
Detoxification mechanisms
Environmental challenge by natural toxins helped to prepare Drosophilae to detox DDT,
Selection
The Drosophila genome is subject to a high degree of selection, especially unusually widespread negative selection compared to other taxa. A majority of the genome is under selection of some sort, and a supermajority of this is occurring in non-coding DNA.[6]
Effective population size has been credibly suggested to positively correlate with the effect size of both negative and positive selection. Recombination is likely to be a significant source of diversity. There is evidence that crossover is positively correlated with polymorphism in D. populations.[6]
Biology
Habitat
Drosophila species are found all around the world, with more species in the tropical regions. Drosophila made their way to the Hawaiian Islands and radiated into over 800 species.[7] They can be found in deserts, tropical rainforest, cities, swamps, and alpine zones. Some northern species hibernate. The northern species D. montana is the best cold-adapted,[8] and is primarily found at high altitudes.[9] Most species breed in various kinds of decaying plant and fungal material, including fruit, bark, slime fluxes, flowers, and mushrooms. The larvae of at least one species, D. suzukii, can also feed in fresh fruit and can sometimes be a pest.[10] A few species have switched to being parasites or predators. Many species can be attracted to baits of fermented bananas or mushrooms, but others are not attracted to any kind of baits. Males may congregate at patches of suitable breeding substrate to compete for the females, or form leks, conducting courtship in an area separate from breeding sites.
Several Drosophila species, including D. melanogaster, D. immigrans, and D. simulans, are closely associated with humans, and are often referred to as domestic species. These and other species (D. subobscura, and from a related genus Zaprionus indianus[11][12][13]) have been accidentally introduced around the world by human activities such as fruit transports.

Reproduction
Males of this genus are known to have the longest sperm cells of any studied organism on Earth, including one species, Drosophila bifurca, that has sperm cells that are 58 mm (2.3 in) long.[14] The cells mostly consist of a long, thread-like tail, and are delivered to the females in tangled coils. The other members of the genus Drosophila also make relatively few giant sperm cells, with that of D. bifurca being the longest.[15] D. melanogaster sperm cells are a more modest 1.8 mm long, although this is still about 35 times longer than a human sperm. Several species in the D. melanogaster species group are known to mate by traumatic insemination.[16]
Drosophila species vary widely in their reproductive capacity. Those such as D. melanogaster that breed in large, relatively rare resources have ovaries that mature 10–20 eggs at a time, so that they can be laid together on one site. Others that breed in more-abundant but less nutritious substrates, such as leaves, may only lay one egg per day. The eggs have one or more respiratory filaments near the anterior end; the tips of these extend above the surface and allow oxygen to reach the embryo. Larvae feed not on the vegetable matter itself, but on the yeasts and microorganisms present on the decaying breeding substrate. Development time varies widely between species (between 7 and more than 60 days) and depends on the environmental factors such as temperature, breeding substrate, and crowding.
Fruit flies lay eggs in response to environmental cycles. Eggs laid at a time (e.g., night) during which likelihood of survival is greater than in eggs laid at other times (e.g., day) yield more larvae than eggs that were laid at those times. Ceteris paribus, the habit of laying eggs at this 'advantageous' time would yield more surviving offspring, and more grandchildren, than the habit of laying eggs during other times. This differential reproductive success would cause D. melanogaster to adapt to environmental cycles, because this behavior has a major reproductive advantage.[17]
Their median lifespan is 35–45 days.[18]
Courtship behavior
The following section is based on the following Drosophila species: Drosophila simulans and Drosophila melanogaster.
Courtship behavior of male Drosophila is an attractive behaviour.[19] Females respond via their perception of the behavior portrayed by the male.[20] Male and female Drosophila use a variety of sensory cues to initiate and assess courtship readiness of a potential mate.[19][20][21] The cues include the following behaviours: positioning, pheromone excretion, following females, making tapping sounds with legs, singing, wing spreading, creating wing vibrations, genitalia licking, bending the stomach, attempt to copulate, and the copulatory act itself.[22][19][20][21] The songs of Drosophila melanogaster and Drosophila simulans have been studied extensively. These luring songs are sinusoidal in nature and varies within and between species.[21]
The courtship behavior of Drosophila melanogaster has also been assessed for sex-related genes, which have been implicated in courtship behavior in both the male and female.[19] Recent experiments explore the role of fruitless (fru) and doublesex (dsx), a group of sex-behaviour linked genes.[23][19]
The fruitless (fru) gene in Drosophila helps regulate the network for male courtship behavior; when a mutation to this gene occurs altered same sex sexual behavior in males is observed.[24] Male Drosophila with the fru mutation direct their courtship towards other males as opposed to typical courtship, which would be directed towards females.[25] Loss of the fru mutation leads back to the typical courtship behavior.[25]
Pheromones
A novel class of pheromones was found to be conserved across the subgenus Drosophila in 11 desert dwelling species.[26] These pheromones are triacylglycerides that are secreted exclusively by males from their ejaculatory bulb and transferred to females during mating. The function of the pheromones is to make the females unattractive to subsequent suitors and thus inhibit courtship by other males.
Polyandry
The following section is based on the following Drosophila species: Drosophila serrata, Drosophila pseudoobscura, Drosophila melanogaster, and Drosophila neotestacea. Polyandry is a prominent mating system among Drosophila.[27][28][29][30] Females mating with multiple sex partners has been a beneficial mating strategy for Drosophila.[27][28][29][30] The benefits include both pre and post copulatory mating. Pre-copulatory strategies are the behaviours associated with mate choice and the genetic contributions, such as production of gametes, that are exhibited by both male and female Drosophila regarding mate choice.[27][28] Post copulatory strategies include sperm competition, mating frequency, and sex-ratio meiotic drive.[27][28][29][30]
These lists are not inclusive. Polyandry among the Drosophila pseudoobscura in North America vary in their number of mating partners.[29] There is a connection between the number of time females choose to mate and chromosomal variants of the third chromosome.[29] It is believed that the presence of the inverted polymorphism is why re-mating by females occurs.[29] The stability of these polymorphisms may be related to the sex-ratio meiotic drive.[30]
However, for Drosophila subobscura, the main mating system is monandry, not normally seen in Drosophila.[31]
Sperm competition
The following section is based on the following Drosophila species: Drosophila melanogaster, Drosophila simulans, and Drosophila mauritiana. Sperm competition is a process that polyandrous Drosophila females use to increase the fitness of their offspring.[32][33][34][35][36] The female Drosophila has two sperm storage organs, the spermathecae and seminal receptacle, that allows her to choose the sperm that will be used to inseminate her eggs.[36] However, some species of Drosophila have evolved to only use one or the other.[37] Females have little control when it comes to cryptic female choice.[35][33] Female Drosophila through cryptic choice, one of several post-copulatory mechanisms, which allows for the detection and expelling of sperm that reduces inbreeding possibilities.[34][33] Manier et al. 2013 has categorized the post copulatory sexual selection of Drosophila melanogaster, Drosophila simulans, and Drosophila mauritiana into the following three stages: insemination, sperm storage, and fertilizable sperm.[35] Among the preceding species there are variations at each stage that play a role in the natural selection process.[35] This sperm competition has been found to be a driving force in the establishment of reproductive isolation during speciation.[38][39]
Laboratory-cultured animals
D. melanogaster is a popular experimental animal because it is easily cultured en masse out of the wild, has a short generation time, and mutant animals are readily obtainable. In 1906, Thomas Hunt Morgan began his work on D. melanogaster and reported his first finding of a white eyed mutant in 1910 to the academic community. He was in search of a model organism to study genetic heredity and required a species that could randomly acquire genetic mutation that would visibly manifest as morphological changes in the adult animal. His work on Drosophila earned him the 1933 Nobel Prize in Medicine for identifying chromosomes as the vector of inheritance for genes. This and other Drosophila species are widely used in studies of genetics, embryogenesis, chronobiology, speciation, neurobiology, and other areas.
However, some species of Drosophila are difficult to culture in the laboratory, often because they breed on a single specific host in the wild. For some, it can be done with particular recipes for rearing media, or by introducing chemicals such as sterols that are found in the natural host; for others, it is (so far) impossible. In some cases, the larvae can develop on normal Drosophila lab medium, but the female will not lay eggs; for these it is often simply a matter of putting in a small piece of the natural host to receive the eggs.[40]
The Drosophila Species Stock Center located at Cornell University in Ithaca, New York, maintains cultures of hundreds of species for researchers.[41]
Use in genetic research
Drosophila is considered one of the most impeccable genetic model organisms - they have furthered genetic research unlike any other model organisms. Both adults and embryos are experimental models.[42] Drosophila is a prime candidate for genetic research because the relationship between human and fruit fly genes is very close.[43] Human and fruit fly genes are so similar, that disease-producing genes in humans can be linked to those in flies. The fly has approximately 15,500 genes on its four chromosomes, whereas humans have about 22,000 genes among their 23 chromosomes. Thus the density of genes per chromosome in Drosophila is higher than the human genome.[44] Low and manageable number of chromosomes make Drosophila species easier to study. These flies also carry genetic information and pass down traits throughout generations, much like their human counterparts. The traits can then be studied through different Drosophila lineages and the findings can be applied to deduce genetic trends in humans. Research conducted on Drosophila help determine the ground rules for transmission of genes in many organisms.[45][4] Drosophila is a useful in vivo tool to analyze Alzheimer's disease.[46] Rhomboid proteases were first detected in Drosophila but then found to be highly conserved across eukaryotes, mitochondria, and bacteria.[47][48] Melanin's ability to protect DNA against ionizing radiation has been most extensively demonstrated in Drosophila, including in the formative study by Hopwood et al 1985.[49]
Microbiome
Like other animals, Drosophila is associated with various bacteria in its gut. The fly gut microbiota or microbiome seems to have a central influence on Drosophila fitness and life history characteristics. The microbiota in the gut of Drosophila represents an active current research field.
Drosophila species also harbour vertically transmitted endosymbionts, such as Wolbachia and Spiroplasma. These endosymbionts can act as reproductive manipulators, such as cytoplasmic incompatibility induced by Wolbachia or male-killing induced by the D. melanogaster Spiroplasma poulsonii (named MSRO). The male-killing factor of the D. melanogaster MSRO strain was discovered in 2018, solving a decades-old mystery of the cause of male-killing. This represents the first bacterial factor that affects eukaryotic cells in a sex-specific fashion, and is the first mechanism identified for male-killing phenotypes.[50] Alternatively, they may protect theirs hosts from infection. Drosophila Wolbachia can reduce viral loads upon infection, and is explored as a mechanism of controlling viral diseases (e.g. Dengue fever) by transferring these Wolbachia to disease-vector mosquitoes.[51] The S. poulsonii strain of Drosophila neotestacea protects its host from parasitic wasps and nematodes using toxins that preferentially attack the parasites instead of the host.[52][53][54]
Predators
Drosophila species are prey for many generalist predators, such as robber flies. In Hawaii, the introduction of yellowjackets from mainland United States has led to the decline of many of the larger species. The larvae are preyed on by other fly larvae, staphylinid beetles, and ants.
Neurochemistry
As with many Eukaryotes, this genus is known to express SNAREs, and as with several others the components of the SNARE complex are known to be somewhat substitutable: Although the loss of SNAP-25 - a component of neuronal SNAREs - is lethal, SNAP-24 can fully replace it. For another example, an R-SNARE not normally found in synapses can substitute for synaptobrevin.[55]
Immunity
The Spätzle protein is a ligand of Toll.[56][57] In addition to melanin's more commonly known roles in the endoskeleton and in neurochemistry, melanization is one step in the immune responses to some pathogens.[56][57] Dudzic et al 2019 additionally find a large number of shared serine protease messengers between Spätzle/Toll and melanization and a large amount of crosstalk between these pathways.[56][57]
Systematics
|

The genus Drosophila as currently defined is paraphyletic (see below) and contains 1,450 described species,[3][58] while the total number of species is estimated at thousands.[59] The majority of the species are members of two subgenera: Drosophila (about 1,100 species) and Sophophora (including D. (S.) melanogaster; around 330 species).
The Hawaiian species of Drosophila (estimated to be more than 500, with roughly 380 species described) are sometimes recognized as a separate genus or subgenus, Idiomyia,[3][60] but this is not widely accepted. About 250 species are part of the genus Scaptomyza, which arose from the Hawaiian Drosophila and later recolonized continental areas.
Evidence from phylogenetic studies suggests these genera arose from within the genus Drosophila:[61][62]
- Liodrosophila Duda, 1922
- Mycodrosophila Oldenburg, 1914
- Samoaia Malloch, 1934
- Scaptomyza Hardy, 1849
- Zaprionus Coquillett, 1901
- Zygothrica Wiedemann, 1830
- Hirtodrosophila Duda, 1923 (position uncertain)
Several of the subgeneric and generic names are based on anagrams of Drosophila, including Dorsilopha, Lordiphosa, Siphlodora, Phloridosa, and Psilodorha.
Genetics
Drosophila species are extensively used as model organisms in genetics (including population genetics), cell biology, biochemistry, and especially developmental biology. Therefore, extensive efforts are made to sequence drosphilid genomes. The genomes of these species have been fully sequenced:[63]
- Drosophila (Sophophora) melanogaster
- Drosophila (Sophophora) simulans
- Drosophila (Sophophora) sechellia
- Drosophila (Sophophora) yakuba
- Drosophila (Sophophora) erecta
- Drosophila (Sophophora) ananassae
- Drosophila (Sophophora) pseudoobscura
- Drosophila (Sophophora) persimilis
- Drosophila (Sophophora) willistoni
- Drosophila (Drosophila) mojavensis
- Drosophila (Drosophila) virilis
- Drosophila (Drosophila) grimshawi
The data have been used for many purposes, including evolutionary genome comparisons. D. simulans and D. sechellia are sister species, and provide viable offspring when crossed, while D. melanogaster and D. simulans produce infertile hybrid offspring. The Drosophila genome is often compared with the genomes of more distantly related species such as the honeybee Apis mellifera or the mosquito Anopheles gambiae.
The modEncode consortium is currently sequencing eight more Drosophila genomes,[64] and even more genomes are being sequenced by the i5K consortium.[65]
Curated data are available at FlyBase.
The Drosophila 12 Genomes Consortium – led by Andrew G. Clark, Michael Eisen, Douglas Smith, Casey Bergman, Brian Oliver, Therese Ann Markow, Thomas Kaufman, Manolis Kellis, William Gelbart, Venky Iyer, Daniel Pollard, Timothy Sackton, Amanda Larracuente, Nadia Singh, and including Wojciech Makalowski, Mohamed Noor, Temple F. Smith, Craig Venter, Peter Keightley, and Leonid Boguslavsky among its contributors – presents ten new genomes and combines those with previously released genomes for D. melanogaster and D. pseudoobscura to analyse the evolutionary history and common genomic structure of the genus. This includes the discovery of transposable elements and illumination of their evolutionary history.[66] Bartolomé et al 2009 find at least 1⁄3 of the TEs in D. melanogaster, D. simulans and D. yakuba have been acquired by horizontal transfer. They find an average of 0.035 HT TEs⁄TE family⁄million years. Bartolomé also finds HT TEs follow other relatedness metrics, with D. melanogaster⇔D. simulans events being twice as common as either of them ⇔ D. yakuba.[66]
See also
- Drosophila hybrid sterility
- Laboratory experiments of speciation
- List of Drosophila species
- Caenorhabditis 'Drosophilae' species supergroup, a group of species generally found on rotten fruits and transported by Drosophila flies
References
- Jones, Daniel (2003) [1917], Peter Roach; James Hartmann; Jane Setter (eds.), English Pronouncing Dictionary, Cambridge: Cambridge University Press, ISBN 978-3-12-539683-8
- "Drosophila". Merriam-Webster Dictionary.
- Gerhard Bächli (1999–2006). "TaxoDros: the database on taxonomy of Drosophilidae".
- Panikker, Priyalakshmi; Xu, Song-Jun; Zhang, Haolin; Sarthi, Jessica; Beaver, Mariah; Sheth, Avni; Akhter, Sunya; Elefant, Felice (9 May 2018). "Restoring Tip60 HAT/HDAC2 Balance in the Neurodegenerative Brain Relieves Epigenetic Transcriptional Repression and Reinstates Cognition". The Journal of Neuroscience. 38 (19): 4569–4583. doi:10.1523/JNEUROSCI.2840-17.2018. PMC 5943982. PMID 29654189.
- Tang, A H; Tu, C P (1994-11-11). "Biochemical characterization of Drosophila glutathione S-transferases D1 and D21". Journal of Biological Chemistry. 269 (45): 27876–27884. doi:10.1016/S0021-9258(18)46868-8. ISSN 0021-9258. PMID 7961718. Retrieved 2021-01-03.
- Hough, Josh; Williamson, Robert J.; Wright, Stephen I. (2013-11-23). "Patterns of Selection in Plant Genomes". Annual Review of Ecology, Evolution, and Systematics. Annual Reviews. 44 (1): 31–49. doi:10.1146/annurev-ecolsys-110512-135851. ISSN 1543-592X.
- Program, U.S. Fish and Wildlife Service/Ecological Services. "Endangered Species | About Us | Featured Species: Relict Leopard Frog". www.fws.gov. Retrieved 2018-03-10.
- Parker, Darren J; Wiberg, R Axel W; Trivedi, Urmi; Tyukmaeva, Venera I; Gharbi, Karim; Butlin, Roger K; Hoikkala, Anneli; Kankare, Maaria; Ritchie, Michael G; Gonzalez, Josefa (August 2018). "Inter and Intraspecific Genomic Divergence in Drosophila montana Shows Evidence for Cold Adaptation". Genome Biology and Evolution. 10 (8): 2086–2101. doi:10.1093/gbe/evy147. PMC 6107330. PMID 30010752.
- Routtu, Jarkko (2007). Genetic and Phenotypic Divergence in Drosophila virilis and D. montana (PDF). Jyväskylä: University of Jyväskylä. p. 13.
- Mark Hoddle. "Spotted Wing Drosophila (Cherry Vinegar Fly) Drosophila suzukii". Center for Invasive Species Research. Retrieved July 29, 2010.
- Vilela, Carlos R (1 January 1999). "Is Zaprionus indianus Gupta, 1970 (Diptera, Drosophilidae) currently colonizing the Neotropical Region". Drosophila Information Service. 82: 37–39.
- van der Linde, Kim; Steck, Gary J.; Hibbard, Ken; Birdsley, Jeffrey S.; Alonso, Linette M.; Houle, David (September 2006). "First records of Zaprionus indianus (Diptera, Drosophilidae), a pest species on commercial fruits, from Panama and the United States of America". Florida Entomologist. 89 (3): 402–404. doi:10.1653/0015-4040(2006)89[402:FROZID]2.0.CO;2. ISSN 0015-4040.
- Castrezana, S. (2007). "New records of Zaprionus indianus Gupta, 1970 (Diptera, Drosophilidae) in North America and a key to identify some Zaprionus species deposited in the Drosophila Tucson Stock Center" (PDF). Drosophila Information Service. 90: 34–36.
- Pitnick, Scott; Spicer, Greg S.; Markow, Therese A. (May 1995). "How long is a giant sperm?". Nature. 375 (6527): 109. Bibcode:1995Natur.375Q.109P. doi:10.1038/375109a0. PMID 7753164. S2CID 4368953.
- Joly, Dominique; Luck, Nathalie; Dejonghe, Béatrice (15 March 2008). "Adaptation to long sperm inDrosophila: correlated development of the sperm roller and sperm packaging". Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 310B (2): 167–178. doi:10.1002/jez.b.21167. PMID 17377954.
- Kamimura, Yoshitaka (2007-08-22). "Twin intromittent organs of Drosophila for traumatic insemination". Biol. Lett. 3 (4): 401–404. doi:10.1098/rsbl.2007.0192. PMC 2391172. PMID 17519186.
- Howlader, Gitanjali; Sharma, Vijay Kumar (August 2006). "Circadian regulation of egg-laying behavior in fruit flies Drosophila melanogaster". Journal of Insect Physiology. 52 (8): 779–785. doi:10.1016/j.jinsphys.2006.05.001. PMID 16781727.
- Broughton, S. J.; Piper, M. D. W.; Ikeya, T.; Bass, T. M.; Jacobson, J.; Driege, Y.; Martinez, P.; Hafen, E.; Withers, D. J.; Leevers, S. J.; Partridge, L. (11 February 2005). "Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands". Proceedings of the National Academy of Sciences. 102 (8): 3105–3110. Bibcode:2005PNAS..102.3105B. doi:10.1073/pnas.0405775102. PMC 549445. PMID 15708981.
- Pan, Y.; Robinett, C. C.; Baker, B. S. (2011). "Turning males on: Activation of male courtship behavior in Drosophila melanogaster". PLOS ONE. 6 (6): e21144. Bibcode:2011PLoSO...621144P. doi:10.1371/journal.pone.0021144. PMC 3120818. PMID 21731661.
- Cook, R. M. (1973). "Courtship Processing in Drosophila melanogaster. II. An Adaptation to Selection for Receptivity to Wingless Males". Animal Behaviour. 21 (2): 349–358. doi:10.1016/S0003-3472(73)80077-6. PMID 4198506.
- Crossley, S. A.; Bennet-Clark, H. C.; Evert, H. T. (1995). "Courtship song components affect male and female Drosophila differently". Animal Behaviour. 50 (3): 827–839. doi:10.1016/0003-3472(95)80142-1. S2CID 53161217.
- Ejima, Aki; Griffith, Leslie C. (October 2007). "Measurement of Courtship Behavior in". Cold Spring Harbor Protocols. 2007 (10): pdb.prot4847. doi:10.1101/pdb.prot4847. PMID 21356948.
- Certel, S. J.; Savella, M. G.; Schlegel, D. C. F.; Kravitz, E. A. (2007). "Modulation of Drosophila male behavioral choice". Proceedings of the National Academy of Sciences. 104 (11): 4706–4711. Bibcode:2007PNAS..104.4706C. doi:10.1073/pnas.0700328104. PMC 1810337. PMID 17360588.
- Demir, Ebru; Dickson, Barry J. (2005-06-03). "fruitless Splicing Specifies Male Courtship Behavior in Drosophila". Cell. 121 (5): 785–794. doi:10.1016/j.cell.2005.04.027. ISSN 0092-8674. PMID 15935764.
- Yamamoto, Daisuke; Kohatsu, Soh (2017-04-03). "What does the fruitless gene tell us about nature vs. nurture in the sex life of Drosophila?". Fly. 11 (2): 139–147. doi:10.1080/19336934.2016.1263778. ISSN 1933-6934. PMC 5406164. PMID 27880074.
- Chin JS, Ellis SR, Pham HT, Blanksby SJ, Mori K, Koh QL, Etges WJ, Yew JY. Sex-specific triacylglycerides are widely conserved in Drosophila and mediate mating behavior. Elife. 2014 Mar 11;3:e01751. doi: 10.7554/eLife.01751. PMID: 24618898; PMCID: PMC3948109
- Frentiu, F. D.; Chenoweth, S. F. (2008). "Polandry and paternity skew in natural and experimental populations of Drosophila serrata". Molecular Ecology. 17 (6): 1589–1596. doi:10.1111/j.1365-294X.2008.03693.x. PMID 18266626. S2CID 23286566.
- Puurtinen, M.; Fromhage, L. (2017). "Evolution of male and female choice in polyandrous systems". Proceedings of the Royal Society B: Biological Sciences. 284 (1851): 20162174. doi:10.1098/rspb.2016.2174. PMC 5378073. PMID 28330914.
- Herrera, P.; Taylor, M. L.; Skeats, A.; Price, T. A. R.; Wedell, N. (2014). "Can patterns of chromosome inversions in Drosophila pseudoobscura predict polyandry across a geographical cline?". Ecology and Evolution. 4 (15): 3072–3081. doi:10.1002/ece3.1165. PMC 4161180. PMID 25247064.
- Pinzone, C. A.; Dyer, K. A. (2013). "Association of polyandry and sex-ratio drive prevalence in natural populations of Drosophila neotestacea". Proceedings: Biological Sciences. 280 (1769): 20131397. doi:10.1098/rspb.2013.1397. PMC 3768301. PMID 24004936.
- Holman, Luke; Freckleton, Robert P.; Snook, Rhonda R. (February 2008). "What use is an infertile sperm? A comparative study of sperm-heteromorphic Drosophila". Evolution. 62 (2): 374–385. doi:10.1111/j.1558-5646.2007.00280.x. PMID 18053077. S2CID 12804737.
- Manier, M. K.; Belote, J. M.; Berben, K. S.; Lüpold, S.; Ala-Honkola, O.; Collins, W. F.; Pitnick, S. (2013). "Rapid Diversification Of Sperm Precedence Traits And Processes Among Three Sibling Drosophila Species". Evolution. 67 (8): 2348–2362. doi:10.1111/evo.12117. PMID 23888856. S2CID 24845539.
- Clark, A. G.; Begun, D. J.; Prout, T. (1999). "Female X Male Interactions in Drosophila Sperm Competition". Science. 238 (5399): 217–220. doi:10.1126/science.283.5399.217. JSTOR 2897403. PMID 9880253. S2CID 43031475.
- Mack, P. D.; Hammock, B. A.; Promislow, D. E. L. (2002). "Sperm competitive ability and genetic relatedness in Drosophila melanogaster: Similarity breeds contempt". Evolution. 56 (9): 1789–1795. doi:10.1111/j.0014-3820.2002.tb00192.x. PMID 12389723. S2CID 2140754.
- Manier, M. K.; Lüpold, S.; Pitnick, S.; Starmer, William T. (2013). "An Analytical Framework for Estimating Fertilization Bias and the Fertilization Set from Multiple Sperm-Storage Organs" (PDF). The American Naturalist. 182 (4): 552–561. doi:10.1086/671782. PMID 24021407. S2CID 10040656.
- Ala-Honkola, O.; Manier, M. K. (2016). "Multiple mechanisms of cryptic female choice act on intraspecific male variation in Drosophila simulans". Behavioral Ecology and Sociobiology. 70 (4): 519–532. doi:10.1007/s00265-016-2069-3. S2CID 17465840.
- Pitnick, Scott; Markow, Therese; Spicer, Greg S. (December 1999). "Evolution of Multiple Kinds of Female Sperm-Storage Organs in Drosophila". Evolution. 53 (6): 1804–1822. doi:10.2307/2640442. ISSN 0014-3820. JSTOR 2640442. PMID 28565462.
- Lüpold, Stefan; Manier, Mollie K.; Puniamoorthy, Nalini; Schoff, Christopher; Starmer, William T.; Luepold, Shannon H. Buckley; Belote, John M.; Pitnick, Scott (2016-05-25). "How sexual selection can drive the evolution of costly sperm ornamentation". Nature. 533 (7604): 535–538. Bibcode:2016Natur.533..535L. doi:10.1038/nature18005. ISSN 0028-0836. PMID 27225128. S2CID 4407752.
- Zajitschek, Susanne; Zajitschek, Felix; Josway, Sarah; Al Shabeeb, Reem; Weiner, Halli; Manier, Mollie K. (2019-05-29). "Costs and benefits of giant sperm and sperm storage organs in Drosophila melanogaster". Journal of Evolutionary Biology. 32 (11): 1300–1309. bioRxiv 10.1101/652248. doi:10.1111/jeb.13529. PMID 31465604. S2CID 191162620.
- "Drosophila Fallén, 1823". www.gbif.org. Retrieved 2022-06-24.
- "The National Drosophila Species Stock Center". College of Agriculture and Life Science, Cornell University.
- Perez-Perri, Joel I.; Noerenberg, Marko; Kamel, Wael; Lenz, Caroline E.; Mohammed, Shabaz; Hentze, Matthias W.; Castello, Alfredo (2020-11-18). "Global analysis of RNA-binding protein dynamics by comparative and enhanced RNA interactome capture". Nature Protocols. Nature Portfolio. 16 (1): 27–60. doi:10.1038/s41596-020-00404-1. ISSN 1754-2189. PMC 7116560. PMID 33208978. (JIPP ORCID: 0000-0001-5395-6549). (MN ORCID: 0000-0002-4834-8888). (SM ORCID: 0000-0003-2640-9560).
- "Why use the fly in research?".
- "ModENCODE | Drosophila as a model organism". Archived from the original on 2020-02-05. Retrieved 2019-11-19.
- Jennings, Barbara H. (May 2011). "Drosophila – a versatile model in biology & medicine". Materials Today. 14 (5): 190–195. doi:10.1016/S1369-7021(11)70113-4.
- Prüßing, K., Voigt, A. & Schulz, J.B. Drosophila melanogaster as a model organism for Alzheimer’s disease. Mol Neurodegeneration 8, 35 (2013). https://doi.org/10.1186/1750-1326-8-35
- Freeman, Matthew (2008). "Rhomboid Proteases and their Biological Functions". Annual Review of Genetics. Annual Reviews. 42 (1): 191–210. doi:10.1146/annurev.genet.42.110807.091628. ISSN 0066-4197. PMID 18605900.
- Freeman, Matthew (2014-10-11). "The Rhomboid-Like Superfamily: Molecular Mechanisms and Biological Roles". Annual Review of Cell and Developmental Biology. Annual Reviews. 30 (1): 235–54. doi:10.1146/annurev-cellbio-100913-012944. ISSN 1081-0706. PMID 25062361. S2CID 31705365.
- Mosse, Irma B.; Dubovic, Boris V.; Plotnikova, Svetlana I.; Kostrova, Ludmila N.; Molophei, Vadim; Subbot, Svetlana T.; Maksimenya, Inna P. (20–25 May 2001). Obelic, B.; Ranogajev-Komor, M.; Miljanic, S.; Krajcar Bronic, I. (eds.). Melanin is Effective Radioprotector against Chronic Irradiation and Low Radiation Doses. IRPA Regional Congress on Radiation Protection in Central Europe: Radiation Protection and Health. INIS. Dubrovnik (Croatia): Croatian Radiation Protection Association. p. 35 (of 268).
{{cite conference}}: CS1 maint: date format (link) - Papageorgiou, Nik (5 July 2018). "Mystery solved: The bacterial protein that kills male fruit flies".
- "Wolbachia". World Mosquito Program.
- Haselkorn, Tamara S.; Jaenike, John (July 2015). "Macroevolutionary persistence of heritable endosymbionts: acquisition, retention and expression of adaptive phenotypes in". Molecular Ecology. 24 (14): 3752–3765. doi:10.1111/mec.13261. PMID 26053523. S2CID 206182327.
- Hamilton, Phineas T.; Peng, Fangni; Boulanger, Martin J.; Perlman, Steve J. (12 January 2016). "A ribosome-inactivating protein in a defensive symbiont". Proceedings of the National Academy of Sciences. 113 (2): 350–355. Bibcode:2016PNAS..113..350H. doi:10.1073/pnas.1518648113. PMC 4720295. PMID 26712000.
- Ballinger, Matthew J.; Perlman, Steve J.; Hurst, Greg (6 July 2017). "Generality of toxins in defensive symbiosis: Ribosome-inactivating proteins and defense against parasitic wasps in Drosophila". PLOS Pathogens. 13 (7): e1006431. doi:10.1371/journal.ppat.1006431. PMC 5500355. PMID 28683136.
- Ungar, Daniel; Hughson, Frederick M. (2003). "SNARE Protein Structure and Function". Annual Review of Cell and Developmental Biology. Annual Reviews. 19 (1): 493–517. doi:10.1146/annurev.cellbio.19.110701.155609. ISSN 1081-0706. PMID 14570579.
- Belmonte, Rebecca L.; Corbally, Mary-Kate; Duneau, David F.; Regan, Jennifer C. (2020-01-31). "Sexual Dimorphisms in Innate Immunity and Responses to Infection in Drosophila melanogaster". Frontiers in Immunology. Frontiers Media. 10: 3075. doi:10.3389/fimmu.2019.03075. ISSN 1664-3224. PMC 7006818. PMID 32076419.
- Cerenius, Lage; Söderhäll, Kenneth (2021). "Immune properties of invertebrate phenoloxidases". Developmental & Comparative Immunology. Elsevier. 122: 104098. doi:10.1016/j.dci.2021.104098. ISSN 0145-305X. PMID 33857469.
- Therese A. Markow; Patrick M. O'Grady (2005). Drosophila: A guide to species identification and use. London: Elsevier. ISBN 978-0-12-473052-6.
- Patterson, Colin (1999). Evolution. Cornell University Press. ISBN 978-0-8014-8594-7.
- Brake, Irina; Bächli, Gerhard (2008). Drosophilidae (Diptera). World Catalogue of Insects. ISBN 978-87-88757-88-0.
- O'Grady, Patrick; DeSalle, Rob (22 February 2008). "Out of Hawaii: the origin and biogeography of the genus Scaptomyza (Diptera: Drosophilidae)". Biology Letters. 4 (2): 195–199. doi:10.1098/rsbl.2007.0575. PMC 2429922. PMID 18296276.
- Remsen, James; O'Grady, Patrick (August 2002). "Phylogeny of Drosophilinae (Diptera: Drosophilidae), with comments on combined analysis and character support". Molecular Phylogenetics and Evolution. 24 (2): 249–264. doi:10.1016/s1055-7903(02)00226-9. PMID 12144760.
- "12 Drosophila Genomes Project". Lawrence Berkeley National Laboratory. Archived from the original on May 27, 2010. Retrieved July 29, 2010.
- "modEncode Comparative Genomics white paper" (PDF). ENCODE. Retrieved December 13, 2013.
- "i5k species nomination summary". Archived from the original on December 15, 2013. Retrieved December 13, 2013.
- Gilbert, Clément; Peccoud, Jean; Cordaux, Richard (2021-01-07). "Transposable Elements and the Evolution of Insects" (PDF). Annual Review of Entomology. Annual Reviews. 66 (1): 355–372. doi:10.1146/annurev-ento-070720-074650. ISSN 0066-4170. PMID 32931312. S2CID 221747772.
Low et al 2007
- Low, Wai Yee; Ng, Hooi Ling; Morton, Craig J.; Parker, Michael W.; Batterham, Philip; Robin, Charles (2007). "Molecular Evolution of Glutathione S-Transferases in the Genus Drosophila". Genetics. Genetics Society of America (OUP). 177 (3): 1363–1375. doi:10.1534/genetics.107.075838. ISSN 0016-6731. PMC 2147980. PMID 18039872.
- Abstract, "We propose..."
- p. 1365, "The lines..."
- p. 1369, "From the..."
External links
- Fly Base FlyBase is a comprehensive database for information on the genetics and molecular biology of Drosophila. It includes data from the Drosophila Genome Projects and data curated from the literature.
- Berkeley Drosophila Genome Project
- Annual Drosophila Research Conference
- AAA: Assembly, Alignment and Annotation of 12 Drosophila species
- UCSC Genome browser
- TaxoDros: The database on Taxonomy of Drosophilidae
- UC San Diego Drosophila Stock Center breeds hundreds of species and supplies them to researchers
- FlyMine Archived 2019-10-04 at the Wayback Machine is an integrated database of genomic, expression and protein data for Drosophila
- The Drosophila Virtual library is library of Drosophila on the web
- Drosophila Melanogaster contains further information.
- C-CAMP Fly facility – In India microinjection service for the generation of transgenic lines, Screening Platforms, Drosophila strain development


.jpg.webp)