RNA splicing, the first stage of post-transcriptional control
Gene expression is the process that transfers genetic information from a gene made of DNA to a functional gene product made of RNA or protein. Genetic Information flows from DNA to RNA by the process of transcription and then from RNA to protein by the process of translation. In order to ensure that the proper products are produced, gene expression is regulated at many different stages during and in between transcription and translation. In eukaryotes, the gene contains extra sequences that do not code for protein. In these organisms, transcription of DNA produces pre-mRNA. These pre-mRNA transcripts often contain regions, called introns, that are intervening sequences which must be removed prior to translation by the process of splicing. The regions of RNA that code for protein are called exons . Splicing can be regulated so that different mRNAs can contain or lack exons, in a process called alternative splicing. Alternative splicing allows more than one protein to be produced from a gene and is an important regulatory step in determining which functional proteins are produced from gene expression. Thus, splicing is the first stage of post-transcriptional control.
Alternative Splicing
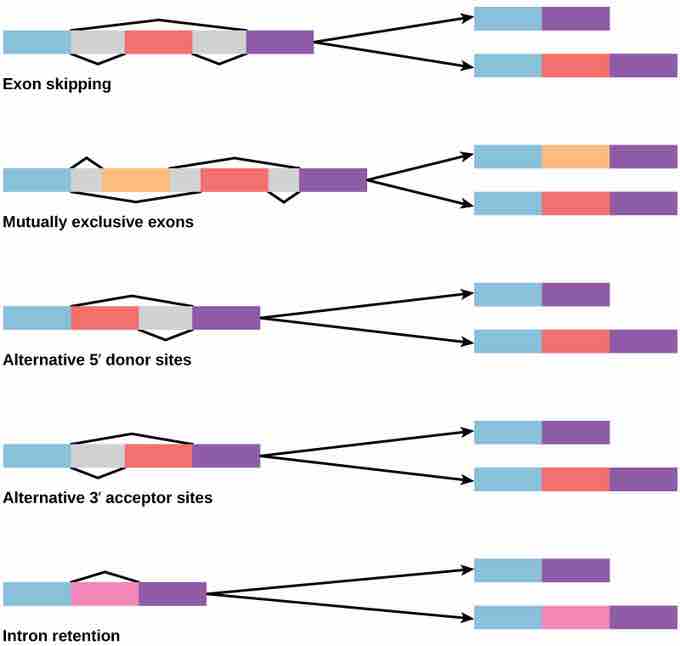
There are five basic modes of alternative splicing.

Alternative Splicing
Pre-mRNA can be alternatively spliced to create different proteins.
Alternative Splicing
Alternative splicing is a process that occurs during gene expression and allows for the production of multiple proteins (protein isoforms) from a single gene coding. Alternative splicing can occur due to the different ways in which an exon can be excluded from or included in the messenger RNA. It can also occur if portions on an exon are excluded/included or if there is an inclusion of introns. For example, if a pre-mRNA has four exons (A, B, C, and D), these can be spliced and translated in a number of different combinations. Exons A, B, and C can be translated together or Exons A, C, and D can be translated. This results in what is called alternative splicing . The pattern of splicing and production of alternatively-spliced messenger RNA is controlled by the binding of regulatory proteins (trans-acting proteins that contain the genes) to cis-acting sites that are found on the pre-RNA. Some of these regulatory proteins include splicing activators (proteins that promote certain splicing sites) and splicing repressors (proteins that reduce the use of certain sites). Some common splicing repressors include: heterogeneous nuclear ribonucleoprotein (hnRNP) and polypyrimidine tract binding protein (PTB). Proteins that are translated from alternatively-spliced messenger RNAs differ in the sequence of their amino acids which results in altered function of the protein. This is one reason why the human genome can encode a wide diversity of proteins. Alternative splicing is a common process that occurs in eukaryotes; most of the multi-exonic genes in humans are spliced alternatively. Unfortunately, abnormal variations in splicing are also the reason why there are many genetic diseases and disorders.

Mechanism of Splicing
Alternative splicing can result in protein isoforms.
Spliceosome
The splicing of messenger RNA is accomplished and catalyzed by a macro-molecule complex known as the spliceosome. The areas for ligation and cleavage are determined by the many sub-units of the spliceosome which include the branch site (A) and the 5' and 3' splice sites. Interactions between these sub-units and the small nuclear ribonucleoproteins (snRNP) found in the spliceosome create a spliceosome A complex which helps determine which introns to leave out and which exons to keep and bind together. Once the introns are cleaved and removed, the exons are joined together by a phosphodiester bond.
Regulatory Proteins
As noted above, splicing is regulated by repressor proteins and activator proteins, which are are also known as trans-acting proteins. Equally as important are the silencers and enhancers that are found on the messenger RNAs, also known as cis-acting sites. These regulatory functions work together in order to create splicing code that determines alternative splicing.