Polymerase Chain Reaction (PCR) is a molecular technique commonly used to amplify nucleic acid sequences. The starting material is a messenger RNA (mRNA) of interest that could be obtained from a wide array of sample types and extracted using commercially available kits and reagents. This mRNA is used to synthesize complementary DNA (cDNA) in a reaction catalyzed by the enzyme reverse transcriptase. The importance of this step is it allows converting a labile RNA into its more stable cDNA form that can be stored and used for multiple applications. The resulting cDNA serves as the template for the PCR reaction. The PCR process can be divided into three steps: DNA denaturation where double-stranded DNA (dsDNA) is separated at temperatures above 90°C, oligonucleotide primers annealing at 50–60°C, and primer extension at 70–78°C. A programmable thermal cycler controls the rate of temperature change, the length of the incubation at each temperature, and the number of times each cycle of temperatures is repeated . The final product of the reaction is called amplicon. It is confirmed by agarose gel electrophoresis for qualitative results.
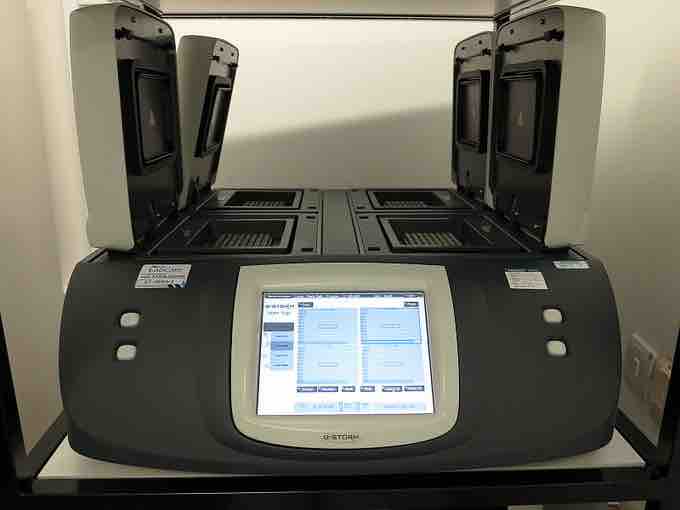
Thermal Cycler
Automated apparatus to amplify DNA sequences using the polymerase chain reaction.
Real-time polymerase chain (RT-PCR) reaction, also called quantitative real-time PCR (qRt-PCR) is used to amplify and quantify targeted DNA molecules. The use of RT-PCR allows for both detection and quantitation of DNA sequences. The quantity can be an absolute number of copies or a relative amount when normalized to DNA input or additional normalizing genes. The procedure for RT-PCR follows the general principles of PCR, but the defining feature is the ability to detect amplified DNA as the reaction progresses in real time.
Real-time PCR can used to amplify low-abundance DNA templates. It is useful in monitoring the accumulating amplicon. Two common methods that are used to product detection in real-time PCR include the use of non-specific flourescent dyes that intercalate with double-stranded DNA or sequence-specific DNA probes that consist of oligonucleotides labeled with a fluorescent reporter (oligoprobes). The fluorescent reporter permits detection after hybridization of the probe with its complementary DNA target . During real-time PCR with oligoprobes, there is a change in signal following direct interaction with the amplicon. The signal is related to the amount of amplicon present during each cycle and will increase as the amount of specific amplicon increases. The detection of amplicon could be visualized on a graph as the amplification progresses. Real-time PCR assays have been extremely useful for studying microbial agents of infectious disease and have proven valuable for basic microbiological research. The ability to amplify templates from a broad selection of specimen has made it an ideal system for application across the various microbiological disciplines.
Real-Time Polymerase Chain Reaction
Real-Time PCR utilizes fluorescent probes to measure DNA amplification.
A new and improved technology called multiplex PCR was introduced to allow the use of one or more primer sets to potentially amplify multiple templates within a single reaction. Up to 20 different reactions can be run simultaneously, therefore lowering the amount of sample used, reducing the reagents consumed, and collecting far more information per reaction, while simplifying data analyses. Multiplex PCR is a challenging application that typically requires more optimization than standard, single amplicon PCR assays. The key to successful multiplex PCR is the ability to define a single set of reaction parameters (reagent concentrations and cycling parameters) that allows for all primers to anneal with high specificity to their target sequences and be extended with the same efficiency. Primer design, as well as the enzyme and buffer system, are critical factors in this challenge. The results from multiplex PCR can be analyzed using gel electrophoresis or using fluorophores for analysis using during the reaction. Ideally, a real-time multiplex PCR should be able to detect, differentiate, and provide a quantitative result for many different targets without a single target influencing the detection of one of the others (cross-talk) and without loss of sensitivity. It is evident that due to the limited number of fluorophoric labels available and the significant overlap in their emission spectra, quantification of multiplex reaction products is often difficult.
Numerous companies have helped overcome this issue by making dyes available that are compatible for use in multiplex PCR. Since its first description in 1988 by Chamberlain et al, this method has been applied in many areas of DNA testing, including analyses of deletions, mutations, and polymorphisms, or quantitative assays and reverse transcription PCR. Typically, it is used for genotyping applications where simultaneous analysis of multiple markers is required, detection of pathogens or genetically modified organisms, or for microsatellite analyses. Multiplex assays can be tedious and time-consuming to establish, requiring lengthy optimization procedures but once optimized numerous high-throughput genomic assays can be achieved at optimum speed.