Replication of Viruses
The genetic material within virus particles and the method by which the material is replicated vary considerably between different types of viruses.
TYPES
DNA viruses: The genome replication of most DNA viruses takes place in the cell's nucleus. If the cell has the appropriate receptor on its surface, these viruses sometimes enter the cell by direct fusion with the cell membrane (e.g., herpesviruses) or, more usually, by receptor-mediated endocytosis. Most DNA viruses are entirely dependent on the host cell's DNA and RNA synthesizing machinery and RNA processing machinery; however, viruses with larger genomes may encode much of this machinery themselves. In eukaryotes the viral genome must cross the cell's nuclear membrane to access this machinery, while in bacteria it need only enter the cell.
RNA viruses: Replication usually takes place in the cytoplasm. RNA viruses can be placed into four different groups, depending on their modes of replication. The polarity of single-stranded RNA viruses largely determines the replicative mechanism, depending on whether or not it can be used directly by ribosomes to make proteins. The other major criterion is whether the genetic material is single-stranded or double-stranded. All RNA viruses use their own RNA replicase enzymes to create copies of their genomes.
Reverse transcribing viruses: These have ssRNA (Retroviridae, Metaviridae, Pseudoviridae) or dsDNA (Caulimoviridae, and Hepadnaviridae) in their particles. Reverse transcribing viruses with RNA genomes (retroviruses), use a DNA intermediate to replicate, whereas those with DNA genomes (pararetroviruses) use an RNA intermediate during genome replication. Both types use a reverse transcriptase, or RNA-dependent DNA polymerase enzyme, to carry out the nucleic acid conversion. Retroviruses integrate the DNA produced by reverse transcription into the host genome as a provirus as a part of the replication process. Pararetroviruses do not, although integrated genome copies, usually of plant pararetroviruses, can give rise to infectious virus. They are susceptible to antiviral drugs that inhibit the reverse transcriptase enzyme, e.g. zidovudine and lamivudine. An example of the first type is HIV, which is a retrovirus. Examples of the second type are the Hepadnaviridae, which includes Hepatitis B virus.
The Baltimore classification developed by David Baltimore is a virus classification system that groups viruses into families, depending on their type of genome (DNA, RNA, single-stranded (ss), double-stranded (ds), etc.) and their method of replication. Classifying viruses according to their genome means that those in a given category will all behave in much the same way, which offers some indication of how to proceed with further research.
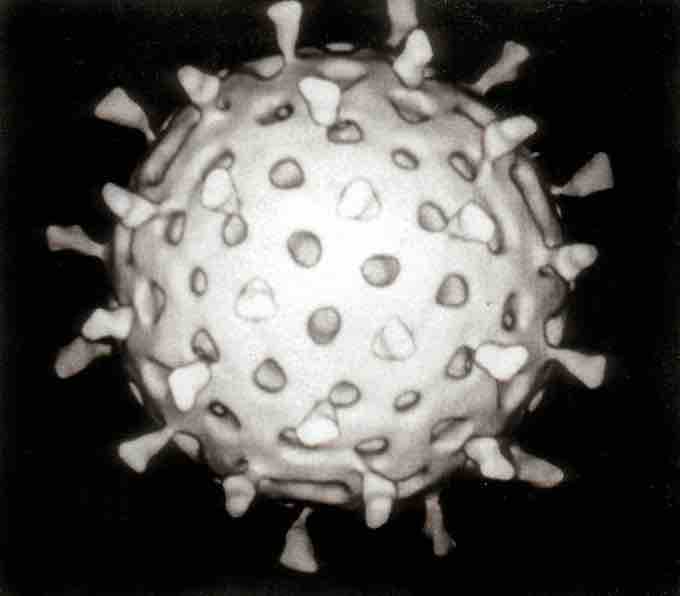
Computer assisted reconstruction of a rotavirus particle
An example of Baltimore Virus classification I: dsDNA virusesII: ssDNA virusesIII: dsRNA virusesIV: (+)ssRNA virusesV: (−)ssRNA virusesVI: ssRNA-RT virusesVII: dsDNA-RT viruses
In summary:
- I: dsDNA viruses (e.g. Adenoviruses, Herpesviruses, Poxviruses)
- II: ssDNA viruses (+)sense DNA (e.g. Parvoviruses)
- III: dsRNA viruses (e.g. Reoviruses)IV: (+)ssRNA viruses (+)sense RNA (e.g. Picornaviruses, Togaviruses)V: (−)ssRNA viruses (−)sense RNA (e.g. Orthomyxoviruses, Rhabdoviruses)
- VI: ssRNA-RT viruses (+)sense RNA with DNA intermediate in life-cycle (e.g. Retroviruses)
- VII: dsDNA-RT viruses (e.g. Hepadnaviruses)