Retroviral RNA Genome
Retroviral genome.
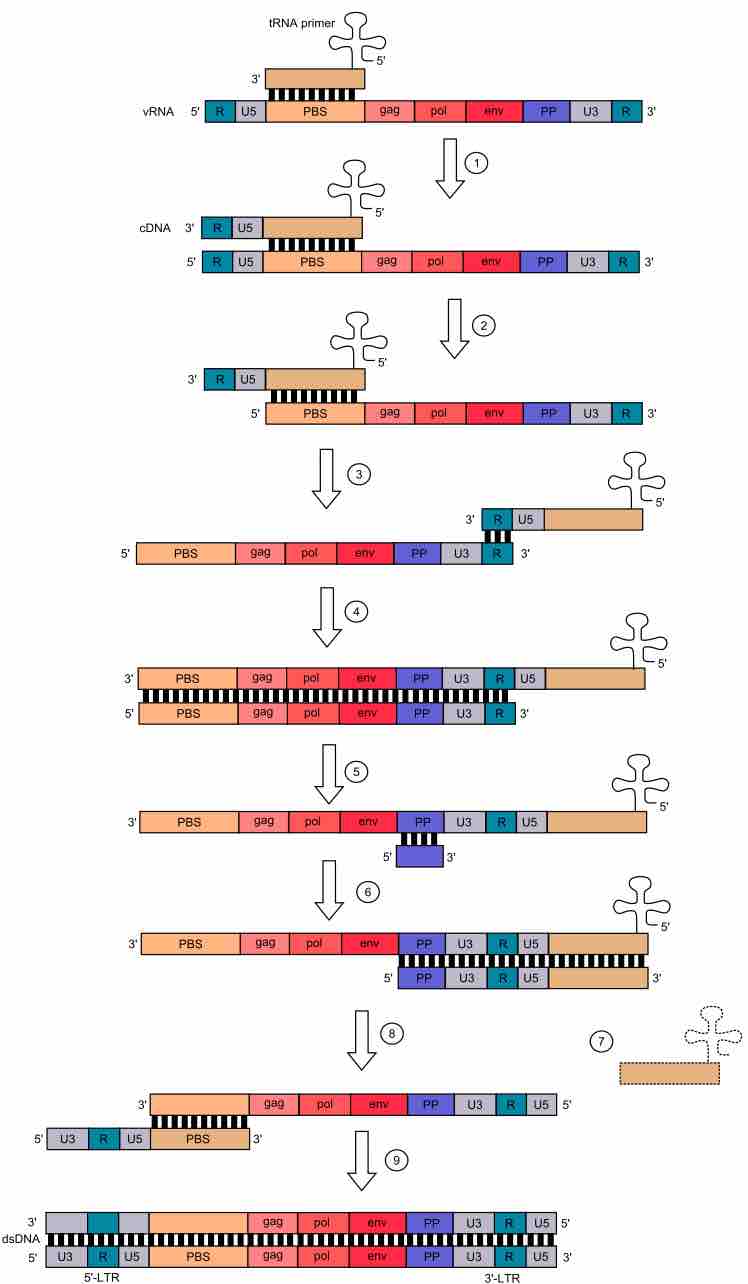
Through the mechanism of reverse transcription by human immunodeficiency virus (HIV), we see what the different genomic elements of a retrovirus are. Reverse transcription occurs in the cytoplasm of host cell. In this process, viral ssRNA is transcribed by the viral reverse transcriptase (RT) into double stranded DNA. Reverse transcription takes place in 5'→3' direction. tRNA ("cloverleaf") hybridizes to PBS and provides -OH group for initiation of reverse transcription. 1) Complementary DNA (cDNA) is formed. 2) Template in RNA:DNA hybrid is degraded by RNase H domain of reverse transcriptase 3) DNA:tRNA is transferred to the 3'-end of the template. 4) First strand synthesis takes place. 5) The rest of viral ssRNA is degraded by RNase H, except for the PP site. 6) Synthesis of second strand of ssDNA is initiated from the 3'-end of the template. tRNA is necessary to synthesis of complementary PBS 7) tRNA is degraded 8) PBS from the second strand hybridizes with the complementary PBS on the first strand. 9) Synthesis of both strands is completed by the DNA Polmerase function of reverse transcriptase. Both dsDNA ends have U3-R-U5 sequences, so called long terminal repeat sequences (3'LTR and 5'LTR, respectively). LTRs mediate integration of the retroviral DNA into another region of the host genome.Key: U3 - promoter region, U5 - recognition site for viral integrase; PBS - primer binding site; PP - polypurine section (polypurine tract); gag, pol, and env. Colors mark complementary sequences.
Source
Boundless vets and curates high-quality, openly licensed content from around the Internet. This particular resource used the following sources: