Replication of viruses primarily involves the multiplication of the viral genome. Replication also involves synthesis of viral messenger RNA (mRNA) from "early" genes (with exceptions for positive sense RNA viruses), viral protein synthesis, possible assembly of viral proteins, then viral genome replication mediated by early or regulatory protein expression. This may be followed, for complex viruses with larger genomes, by one or more further rounds of mRNA synthesis: "late" gene expression is, in general, necessary for structural or virion proteins. Viral replication usually takes place in the cytoplasm .
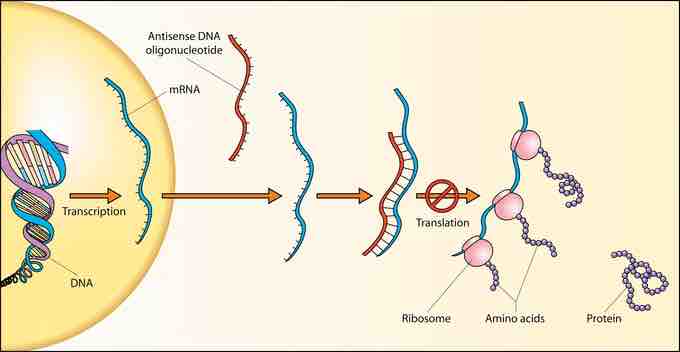
DNA to RNA to protein
Schematic showing how antisense DNA strands can interfere with protein translation.
Viruses that replicate via RNA intermediates need an RNA-dependent RNA-polymerase to replicate their RNA, but animal cells do not seem to possess a suitable enzyme. Therefore, this type of animal RNA virus needs to code for an RNA-dependent RNA polymerase. No viral proteins can be made until viral messenger RNA is available; thus, the nature of the RNA in the virion affects the strategy of the virus: In plus-stranded RNA viruses, the virion (genomic) RNA is the same sense as mRNA and so functions as mRNA. This mRNA can be translated immediately upon infection of the host cell . Examples: poliovirus (picornavirus), togaviruses, and flaviviruses.
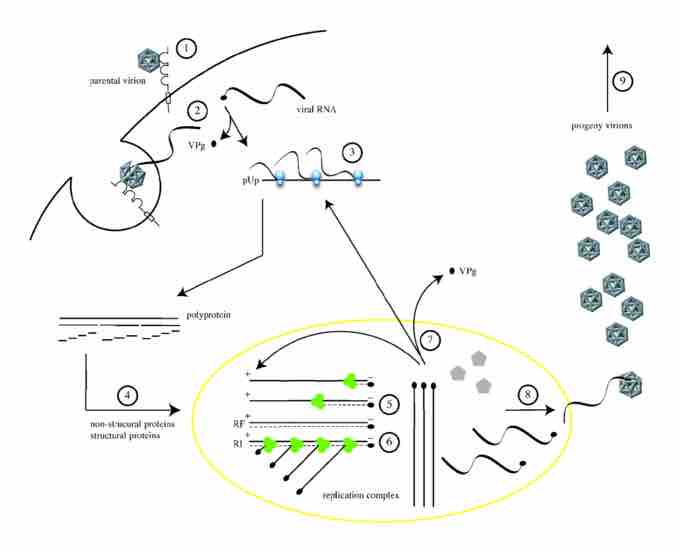
The replication cycle of poliovirus
The cellular life cycle of poliovirus. It is initiated by binding of a poliovirion to the cell surface macromolecule CD155, which functions as the receptor (1). Uncoating of the viral RNA is mediated by receptor-dependent destabilization of the virus capsid (2). Cleavage of the viral protein VPg is performed by a cellular phosphodiesterase, and translation of the viral RNA occurs by a cap-independent (IRES-mediated) mechanism (3). Proteolytic processing of the viral polyprotein yields mature structural and non-structural proteins (4). The positive-sense RNA serves as template for complementary negative-strand synthesis, thereby producing a double-stranded RNA (replicative form, RF) (5). Initiation of many positive strands from a single negative strand produces the partially single-stranded replicative intermediate (RI) (6). The newly synthesized positive-sense RNA molecules can serve as templates for translation (7) or associate with capsid precursors to undergo encapsidation and induce the maturation cleavage of VP0 (8), which ultimately generates progeny virions. Lysis of the infected cell results in release of infectious progeny virions (9).
RNA viruses are classified into distinct groups depending on their genome and mode of replication (and the numerical groups based on the older Baltimore classification).
Positive-sense ssRNA viruses (Group IV) have their genome directly utilized as if it were mRNA, with host ribosomes translating it into a single protein which is modified by host and viral proteins to form the various proteins needed for replication. One of these includes RNA-dependent RNA polymerase (RNA replicase), which copies the viral RNA to form a double-stranded replicative form, in turn this directs the formation of new virions.