Automated patch clamp
Automated patch clamping is beginning[1] to replace manual patch clamping as a method to measure the electrical activity of individual cells. Different techniques are used to automate patch clamp recordings from cells in cell culture and in vivo. This work has been ongoing since the late 1990s by research labs and companies trying to reduce its complexity and cost of patch clamping manually. Patch clamping for a long time was considered an art form and is still very time consuming and tedious, especially in vivo. The automation techniques try to reduce user error and variability in obtaining quality electrophysiology recordings from single cells.
Manual patch clamp
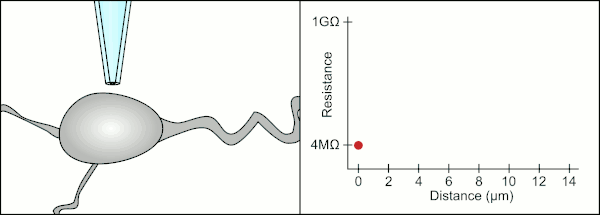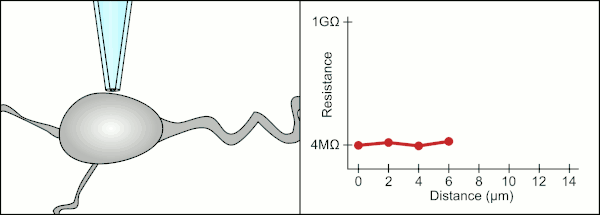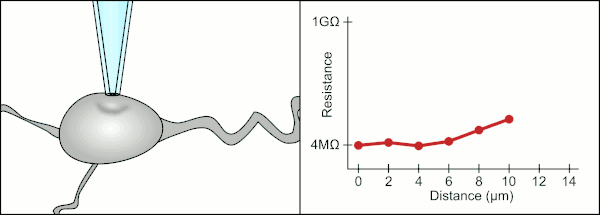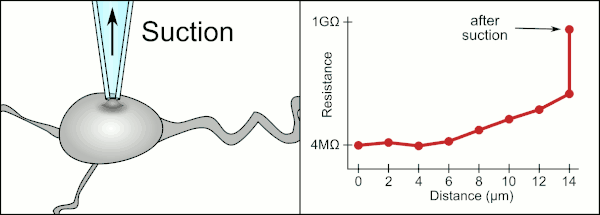
The traditional manual method to patch clamp using glass pipettes was developed by Erwin Neher and Bert Sakmann and required a highly skilled technician. The technician would position the glass pipette near a cell and apply the appropriate suction to create an electrical seal between the pipette and the cell membrane. This seal ensures a quality recording by preventing any current from leaking out between the tip of the pipette and the cell membrane. This seal is made when the membrane of the cell chemically binds with the tip of the pipette so that the inside of the pipette is only connected to the cytoplasm of the cell. This membrane-glass connection or seal is called a "gigaseal".[2]
The technician traditionally used their mouth to provide the precise pressures required to seal it to the cell. In addition to controlling the pressure, the technician must also position the pipette at precisely the correct distance from the cell so that the membrane will seal with it. Using a micromanipulator, the pipette is moved towards the cell until the technician sees a change in the electrical resistance between the fluid inside of the pipette and the surrounding fluid (see animation). This typically requires 3–12 months of training before a technician is able to reliably record from cells. The technician is essentially performing a balancing act trying to watch and manipulate several systems simultaneously (motion, pressure, and electrical signals). Unless each portion of the process is performed accurately and with the right timing, the seal will not be formed properly and the technician will have to replace the pipette and start over.
These challenges reduce the number of recordings a technician can obtain, and significantly increase the cost. Automation seeks to reduce the time, complexity and cost of manual patch clamping. Improving throughput will also be key to enabling high throughput patch-seq for simultaneously combining electrophysiology, morphology, and transcriptomic properties of neurons at the scale comparable to other sequencing methods[3]
Automation systems
The automation technique varies, depending on the surrounding environment of the cells. For cells in vivo, this typically means that the cells are in the brain and surrounded by other cells. This environment also contains blood vessels, dendrites, axons, and glial cells which make it harder to form a gigaseal by clogging the 1-2μm diameter pipette tip. Here, the precise control of pressure and position at the pipette tip plays a big role in preventing clogging, and detecting whether a cell is near the tip of the pipette, as discussed above.
Cells in vitro can be suspended in a fluid, made to adhere to a culture dish, or remain part of a piece of tissue that has been removed from the animal. These environments typically don't have to compensate for motion of the tissue due to the heartbeat or breathing of an animal. In the case of cells in suspension, the pipette is completely replaced with a microchip with holes that can create gigaseals and measure the electrical activity. Clogging is also less problematic for cells or tissue in culture dishes because the cells and pipette can be seen through a microscope which helps the technician avoid everything but the cell of interest.
Each of these automated systems must perform several tasks. It must position the cell next to the tip of a pipette, or some other device with a 1-2μm hole, control the pressure at the hole, and control the voltage inside the cell.
In vivo
One example of in vivo patch clamping was shown by Kodandaramaiah, et al.[4] In this case the pressure control consisted of a set of electronic valves and electronic pressure regulators to provide three pressures that were previously provided by a technician (high pressure 800-1000mbar, low pressure 20-30mbar, and a small vacuum 15-150mbar). Three electronic valves switched between the three pressures and atmospheric pressure. The high pressure was used to prevent clogging the pipette, the low pressure was used when searching for cells, and the vacuum was used to help the gigasealing process. These were all controlled by a computer, to select among the pressures as the resistance at the tip of the pipette changed.
The manual position control in this case was replaced by a computer controlled piezoelectric micromanipulator that moved the pipette in discrete 2-3μm steps into the tissue until it made contact with a cell. This precision control is much more accurate and repeatable than manual positioning and doesn't require an operator.
The computer also calculates and tracks the change in the electrical resistance as the pipette makes contact with the cell. It sends a voltage signal in the form of a square wave down the pipette which either exits the end of the pipette or is blocked by the cell membrane. When the membrane blocks it, the computer stops the motion of the pipette and applies suction to form the gigaseal. This automation eliminates the decision-making a technician had to perform, and unlike a technician, the computer can perform these tasks tirelessly and with greater precision.
All of these steps are performed in the same logical sequence as manual patch clamping, but don't require extensive training to perform, and are completely controlled by the computer. This reduces the expense required to obtain patch clamp recordings and increases the repeatability and robustness of recording in the living brain.
In suspension
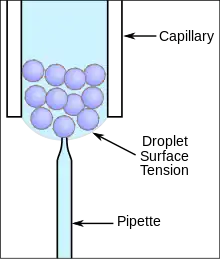
Many types of systems have been developed for patch clamping cells in suspension cultures. One system uses a traditional pipette and cells in a droplet suspension culture to obtain patch clamp recordings (see figure). This has the added benefit of using traditional pipette fabrication systems that heat a glass capillary and pull it lengthwise to create the tapered tip used in patch clamping.
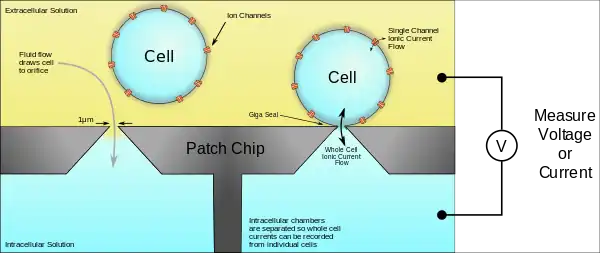
More common automation systems for suspensions cultures use microchips with tiny (1-2μm) holes in a planar substrate instead of pipettes to create the gigaseal and record from single cells. Patch chips were developed in the early 2000s as a result of the improvement of microfabrication technologies developed by the semiconductor industry. Chips are typically made from silicon, glass, PDMS, polymide. The patch chip systems are usually more complex and expensive but have the added benefit of parallel and hands-free operation.[5][6][7]
Ordinarily, neurons don't grow in suspension cultures, but other cell types can. Some can be transfected with genes to create the membrane ion channels of interest. This means that a cell that normally doesn't have electrical activity can grow ion channels in its membrane that will generate ionic currents. Because the cells are dissociated from one another in suspension cultures, the ionic currents in a single cell can be measured with precision. This allows researchers to study ion channel behavior in more controlled environments without currents from other cells interfering, as usually occurs in neural networks. This is particularly useful in drug screening studies where the target is a specific protein.[8] Because handling cells in suspension is much easier than handling cells in culture or in vivo, patch clamp recordings can be obtained much faster and more reliably this way, which increases productivity, making the screening of thousands of compounds possible.[9]
Neurons derived from stem cells cultured adherently can be lifted into suspension and have been successfully used on planar patch clamp devices.[10] Ion channels such as voltage-gated sodium channels, voltage-gated potassium channels and ionotropic ligand-gated ion channels opened by the ligand GABA were recorded from these cells using automated and manual patch clamp.[11]
In culture
There are many in vitro methods for the automated patch clamping of cultured cells or slices of brain tissue.
One uses a patch chip, like those discussed above, along with surface treatments that cause the cultured cells to migrate to the orifices where the gigaseal is formed as they grow.[12] By allowing the neurons to grow in culture, they form networks spontaneously, like those in the brain, which is more like the natural tissues than isolated cells in suspension.
In another method, cells are removed from an animal and cultured on the patch chip for 2–4 hours as they spontaneously form gigaseals with polyimide and PDMS patch chips [13] This system requires no external equipment to form gigaseals.
Another technique automates the positioning of patch clamping cells in cultures. It uses a nanopipette on a precise, piezo-actuated stage to scan a surface within a culture dish. As it scans, it maintains a constant electrical capacitance between the tip of the pipette and the surface or cells beneath it by moving it up and down. (As it moves close to a cell, the capacitance increases, so the actuator moves the pipette away, and vice versa.) This give a precise topographical mapping of the surface within the culture dish. After the cells have been mapped, the computer moves the pipette over to a selected cell and lowers it to form a gigaseal with it.[14]
Another technique simply automates the business of carefully making contact with cells. The operator positions a pipette over the sample and then lets the automated software take over, lowering the pipette and seeking to detect an increase in resistance on the pipette as it makes contact with a cell. At this point the process ends, and a technician creates the gigaseal manually.
Development and acceptance
Patch-clamp automation instrumentation became commercially available in 2003. Due to the initial high cost this near to 20 years old technology was originally intended to serve the biotech and pharmaceutical industries but for the last years its presence has been growing in academia and nonprofit settings, given its increasing proven technical reliability and relative accessibility in cost. A growing number of universities and other academic institutions now have laboratories and core facilities equipped with patch-clamp automated apparatuses in connection and coexisting with other associated or complementary technologies and methods.[15] The acceptance and recognition of automation patch-clamp electrophysiology is reflected in the exponential growth of scientific literature published with results obtained with this revolutionary new technology [16]
References
- ↑ Wood C, Williams C, Waldron GJ (May 2004). "Patch clamping by numbers". Drug Discovery Today. 9 (10): 434–41. doi:10.1016/S1359-6446(04)03064-8. PMID 15109948.
- ↑ Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ (August 1981). "Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches". Pflügers Archiv. 391 (2): 85–100. CiteSeerX 10.1.1.456.107. doi:10.1007/BF00656997. PMID 6270629. S2CID 12014433.
- ↑ Tripathy, Shreejoy J.; Toker, Lilah; Bomkamp, Claire; Mancarci, B. Ogan; Belmadani, Manuel; Pavlidis, Paul (2018-10-08). "Assessing Transcriptome Quality in Patch-Seq Datasets". Frontiers in Molecular Neuroscience. 11: 363. doi:10.3389/fnmol.2018.00363. ISSN 1662-5099. PMC 6187980. PMID 30349457.
- ↑ Kodandaramaiah SB, Franzesi GT, Chow BY, Boyden ES, Forest CR (June 2012). "Automated whole-cell patch-clamp electrophysiology of neurons in vivo". Nature Methods. 9 (6): 585–7. doi:10.1038/nmeth.1993. PMC 3427788. PMID 22561988.
- ↑ Jones KA, Garbati N, Zhang HM, Large CH (2009). "Automated Patch Clamping Using the QPatch". High Throughput Screening. Methods in Molecular Biology. Vol. 565. pp. 209–23. doi:10.1007/978-1-60327-258-2_10. ISBN 978-1-60327-257-5. PMID 19551364.
- ↑ Chen CY, Tu TY, Chen CH, Jong DS, Wo AM (August 2009). "Patch clamping on plane glass-fabrication of hourglass aperture and high-yield ion channel recording". Lab on a Chip. 9 (16): 2370–80. doi:10.1039/B901025D. PMID 19636469.
- ↑ Obergrussberger A, Stölzle-Feix S, Becker N, Brüggemann A, Fertig N, Möller C (2015). "Novel screening techniques for ion channel targeting drugs". Channels. 9 (6): 367–75. doi:10.1080/19336950.2015.1079675. PMC 4850050. PMID 26556400.
- ↑ Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R (April 2008). "High-throughput electrophysiology: an emerging paradigm for ion-channel screening and physiology". Nature Reviews. Drug Discovery. 7 (4): 358–68. doi:10.1038/nrd2552. PMID 18356919. S2CID 9294213.
- ↑ Obergrussberger A, Brüggemann A, Goetze TA, Rapedius M, Haarmann C, Rinke I, et al. (December 2016). "Automated Patch Clamp Meets High-Throughput Screening: 384 Cells Recorded in Parallel on a Planar Patch Clamp Module". Journal of Laboratory Automation. 21 (6): 779–793. doi:10.1177/2211068215623209. PMID 26702021.
- ↑ Haythornthwaite A, Stoelzle S, Hasler A, Kiss A, Mosbacher J, George M, et al. (October 2012). "Characterizing human ion channels in induced pluripotent stem cell-derived neurons". Journal of Biomolecular Screening. 17 (9): 1264–72. doi:10.1177/1087057112457821. PMID 22923790.
- ↑ Haythornthwaite A, Stoelzle S, Hasler A, Kiss A, Mosbacher J, George M, et al. (October 2012). "Characterizing human ion channels in induced pluripotent stem cell-derived neurons". Journal of Biomolecular Screening. 17 (9): 1264–72. doi:10.1177/1087057112457821. PMID 22923790.
- ↑ Py C, Martina M, Diaz-Quijada GA, Luk CC, Martinez D, Denhoff MW, et al. (2011). "From understanding cellular function to novel drug discovery: the role of planar patch-clamp array chip technology". Frontiers in Pharmacology. 2: 51. doi:10.3389/fphar.2011.00051. PMC 3184600. PMID 22007170.
- ↑ Martinez D, Py C, Denhoff MW, Martina M, Monette R, Comas T, et al. (December 2010). "High-fidelity patch-clamp recordings from neurons cultured on a polymer microchip". Biomedical Microdevices. 12 (6): 977–85. CiteSeerX 10.1.1.362.9850. doi:10.1007/s10544-010-9452-z. PMID 20694518. S2CID 14441945.
- ↑ Gorelik J, Gu Y, Spohr HA, Shevchuk AI, Lab MJ, Harding SE, et al. (December 2002). "Ion channels in small cells and subcellular structures can be studied with a smart patch-clamp system". Biophysical Journal. 83 (6): 3296–303. Bibcode:2002BpJ....83.3296G. doi:10.1016/S0006-3495(02)75330-7. PMC 1302405. PMID 12496097.
- ↑ Picones A, Loza-Huerta A, Segura-Chama P, Lara-Figueroa CO (2016). "Contribution of Automated Technologies to Ion Channel Drug Discovery". Advances in Protein Chemistry and Structural Biology. 104: 357–378. doi:10.1016/bs.apcsb.2016.01.002. ISBN 9780128048269. PMID 27038379.
- ↑ Picones A (2015). "Ion channels as medicinal targets of biological toxins: the impact of automated patch-clamp electrophysiology". Current Topics in Medicinal Chemistry. 15 (7): 631–7. doi:10.2174/1568026615666150309145928. PMID 25751269.