Cortical thymic epithelial cells
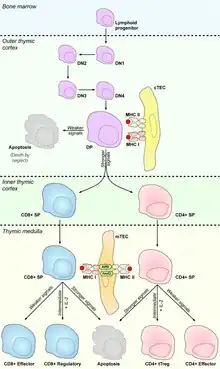
Cortical thymic epithelial cells (cTECs) form unique parenchyma cell population of the thymus which critically contribute to the development of T cells.
Thymus tissue is compartmentalized into cortex and medulla and each of these two compartments comprises its specific thymic epithelial cell subset. cTECs reside in the outer part- cortex, which mostly serves as a developmental site for T cells. Precursors of T cells originate in the bone marrow from which they migrate via bloodstream into thymic cortex, where they encounter stromal cells including cTECs, which form the microenvironment crucial for proliferation and development of T cells by expression of DLL4 (delta-like notch ligand 4), cytokines IL-7, TGFβ or stem cell factor and chemokines CCL25, CXCL12 or CCRL1 etc.[1] Essential part of T cell development forms process called VDJ recombination, mediated by RAG recombinases, that stochastically changes DNA sequences of T cell receptors (TCR) and endows them with diverse recognition specificity. Thanks to this process, T cells can recognize vast repertoire of pathogens, but also self-peptides or even their TCRs don't respond to any surrounding signals. Major role of thymic epithelial cells is to test, whether TCRs are "functional" and on the other hand "harmless" to our body. While cTECs control the functionality of TCRs during the process called positive selection, Medullary thymic epithelial cells (mTECs) that home in the inner part of the thymus- medulla, present on their MHC molecules self-peptides, generated mostly by protein Autoimmune regulator, to eliminate T cells with self-reactive TCRs via processes of central tolerance e.g. negative selection and protect the body against development of autoimmunity.[2]
Positive selection of T cells
Major function of cTECs is to positively select those T cells that are capable to recognize and interact with MHC molecules on their surface [3]. Once T cell precursors enter the thymic cortex, they start their transformation from double negative stages (T cell without surface expression of CD4 and CD8 co-receptors) to a double positive stage (T cell with surface expression of both co-receptors) that expresses fully recombined TCR.[4] This stage undergoes above mentioned selection process.[5]
Double positive–single positive transition
Interaction between TCR of double positive T cell and MHC I molecule leads to loss of CD4 expression and double positive T cell becomes CD8 single positive T cell, conversely, engagement of MHC II molecule leads to the development into CD4 single positive T cell.[6] It was also described that CD8/CD4 restriction is influenced by transcription factors Runx3, in the case of CD8 restriction,[7] and Th-POK[8] which directs the development into CD4 T cell lineage and represses the expression of Runx3.[9] More than 90% of double positive T cells are unable to reach this interaction and they die by neglect.[10]
Cortex–medulla migration
Besides double positive-single positive transition, TCR-MHC interaction also triggers the expression of CCR7, chemokine receptor which recognizes chemokines CCL19 and CCL21, that are largely produced by mTECs in the medulla, and positively selected T cells start to migrate to medulla via their gradient.[11][12]
Unique proteolytic pathways
It is incompletely understood whether presence of peptide ligands on MHC molecules of cTECs plays some role in positive selection. But it is likely that these peptide-MHC complexes are unique and different from self-peptides presented by mTECs, since cTECs developed unique proteolytic pathways. Indeed, there is slight evidence focused on unique cTEC peptide ligands,[13][14][15] nevertheless, its more systematic characterization is still required.
Thymoproteasome (β5t)
Enzymatic machinery for MHC I antigen processing and presentation in cTECs involves thymoproteasome, which is defined by the presence of β5t subunit encoded by Psmb11 gene.[16] Knockout of this gene revealed only slight reduction in positive selection of CD8 T cells, but TCR repertoire of these cells was shown to be limited [17] and they revealed impaired immunological properties e.g. bad antigen responsiveness and failure to maintain naive population in the periphery.[18] β5t subunit was shown to reduce chymotrypsin-like activity of thymoproteasomes, resulting in generation of low affinity peptides.[16] Such finding was confirmed by study that was focused on properties of thymoproteasome- chopped peptides.[15] Importantly, low affinity interactions are considered to result in positive selection, whereas high affinity interactions are typical for negative selection and interaction with mTECs.[3]
Cathepsin L
MHC II processing and presentation in cTECs took advantage of several proteolytic pathways including cathepsin L, encoded by Ctsl gene. Cathepsin S which is produced by most of the antigen- presenting cells along with mTECs is absent in cTECs.[19] Cathepsin L not only cleaves invariant chain as other cathepsins, nevertheless was shown to cleave peptides for MHC II presentation and enlarge the pool of cTEC unique peptide ligands.[20] Ctsl knockout mouse revealed severe reduction in frequency and repertoire of CD4 T cells and impairment of invariant chain degradation.[19] Another study revealed that reduction of T cell repertoire wasn't caused by absence of invariant chain degradation, rather due to alterations in repertoire of cathepsin L cleaved peptides.[20]
Thymus specific serine protease
Thymus specific serine protease is another cTEC specific enzyme, encoded by Prss16 gene, which is also involved in MHC II peptide processing.[21] Prss16 knockout mice revealed reduced repertoire of positively selected CD4 T cells.[22]
Macroautophagy
Common feature of cTECs and mTECs is constitutive macroautophagy.[23] This process involves engulfment of portion of cytoplasm that contains organelles and vesicles into autophagosome that fuses with late endosomes or lysosomes and its content is chopped to small peptides.[24] cTECs and mTECs utilize this endogenous pathway for MHC II presentation during selection processes, instead of common loading of exogenous peptides. Mouse with deficient macroautophagy, specifically in the thymus, revealed reduced numbers and repertoire of CD4 T cells.[25]
Development
cTECs and mTECs originate from endoderm, more specifically from the third pharyngeal pouch[26] and it has been shown that they share common progenitor cell.[27][28] Importantly, mTECs during their development possess classical markers of cTECs including CD205[29] and β5t [30] which are completely absent in mature mTECs,[31] suggesting another possible cTEC function, namely they might serve as a progenitor cell reservoir for mTECs. Indeed, several lineage tracing studies confirmed that cTEC progenitors [32] or even mature cTECs [33][34] are capable to give rise to mTECs.
Nevertheless, there is available series of publications which suggests different mTEC progenitor pools [35][36] or even argue that cTECs and mTECs reveal distinct unipotent progenitor cells.[37][38]
References
- ↑ Ohigashi, Izumi; Kozai, Mina; Takahama, Yousuke (2016-04-18). "Development and developmental potential of cortical thymic epithelial cells". Immunological Reviews. 271 (1): 10–22. doi:10.1111/imr.12404. ISSN 0105-2896. PMID 27088904. S2CID 205213554.
- ↑ Klein, Ludger; Hinterberger, Maria; Wirnsberger, Gerald; Kyewski, Bruno (December 2009). "Antigen presentation in the thymus for positive selection and central tolerance induction". Nature Reviews Immunology. 9 (12): 833–844. doi:10.1038/nri2669. ISSN 1474-1733. PMID 19935803. S2CID 205490965.
- 1 2 Klein, Ludger; Kyewski, Bruno; Allen, Paul M.; Hogquist, Kristin A. (2014-05-16). "Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see)". Nature Reviews Immunology. 14 (6): 377–391. doi:10.1038/nri3667. ISSN 1474-1733. PMC 4757912. PMID 24830344.
- ↑ Petrie, Howard T. (November 2002). "Role of thymic organ structure and stromal composition in steady-state postnatal T-cell production". Immunological Reviews. 189: 8–19. doi:10.1034/j.1600-065X.2002.18902.x. ISSN 0105-2896. PMID 12445261. S2CID 34924894.
- ↑ Starr, Timothy K.; Jameson, Stephen C.; Hogquist, Kristin A. (April 2003). "Positive Andnegativeselection Oft Cells". Annual Review of Immunology. 21 (1): 139–176. doi:10.1146/annurev.immunol.21.120601.141107. ISSN 0732-0582. PMID 12414722.
- ↑ Germain, Ronald N. (May 2002). "T-cell development and the CD4–CD8 lineage decision". Nature Reviews Immunology. 2 (5): 309–322. doi:10.1038/nri798. ISSN 1474-1733. PMID 12033737. S2CID 21479833.
- ↑ Setoguchi, Ruka; Tachibana, Masashi; Naoe, Yoshinori; Muroi, Sawako; Akiyama, Kaori; Tezuka, Chieko; Okuda, Tsukasa; Taniuchi, Ichiro (2008-02-08). "Repression of the Transcription Factor Th-POK by Runx Complexes in Cytotoxic T Cell Development". Science. 319 (5864): 822–825. Bibcode:2008Sci...319..822S. doi:10.1126/science.1151844. ISSN 0036-8075. PMID 18258917. S2CID 6695125.
- ↑ He, Xiao; He, Xi; Dave, Vibhuti P.; Zhang, Yi; Hua, Xiang; Nicolas, Emmanuelle; Xu, Weihong; Roe, Bruce A.; Kappes, Dietmar J. (February 2005). "The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment". Nature. 433 (7028): 826–833. Bibcode:2005Natur.433..826H. doi:10.1038/nature03338. ISSN 0028-0836. PMID 15729333.
- ↑ Luckey, Megan A; Kimura, Motoko Y; Waickman, Adam T; Feigenbaum, Lionel; Singer, Alfred; Park, Jung-Hyun (2014-06-01). "The transcription factor ThPOK suppresses Runx3 and imposes CD4+ lineage fate by inducing the SOCS suppressors of cytokine signaling". Nature Immunology. 15 (7): 638–645. doi:10.1038/ni.2917. ISSN 1529-2908. PMC 6693509. PMID 24880459.
- ↑ Palmer, Ed (May 2003). "Negative selection — clearing out the bad apples from the T-cell repertoire". Nature Reviews Immunology. 3 (5): 383–391. doi:10.1038/nri1085. ISSN 1474-1733. PMID 12766760. S2CID 28321309.
- ↑ Ueno, Tomoo; Saito, Fumi; Gray, Daniel H. D.; Kuse, Sachiyo; Hieshima, Kunio; Nakano, Hideki; Kakiuchi, Terutaka; Lipp, Martin; Boyd, Richard L. (2004-08-16). "CCR7 Signals Are Essential for Cortex–Medulla Migration of Developing Thymocytes". Journal of Experimental Medicine. 200 (4): 493–505. doi:10.1084/jem.20040643. ISSN 0022-1007. PMC 2211934. PMID 15302902.
- ↑ Kurobe, Hirotsugu; Liu, Cunlan; Ueno, Tomoo; Saito, Fumi; Ohigashi, Izumi; Seach, Natalie; Arakaki, Rieko; Hayashi, Yoshio; Kitagawa, Tetsuya (February 2006). "CCR7-Dependent Cortex-to-Medulla Migration of Positively Selected Thymocytes Is Essential for Establishing Central Tolerance". Immunity. 24 (2): 165–177. doi:10.1016/j.immuni.2005.12.011. ISSN 1074-7613. PMID 16473829.
- ↑ Lo, Wan-Lin; Felix, Nathan J; Walters, James J; Rohrs, Henry; Gross, Michael L; Allen, Paul M (2009-10-04). "An endogenous peptide positively selects and augments the activation and survival of peripheral CD4+ T cells". Nature Immunology. 10 (11): 1155–1161. doi:10.1038/ni.1796. ISSN 1529-2908. PMC 2764840. PMID 19801984.
- ↑ Santori, Fabio R.; Kieper, William C.; Brown, Stuart M.; Lu, Yun; Neubert, Thomas A.; Johnson, Kenneth L.; Naylor, Stephen; Vukmanović, Stanislav; Hogquist, Kristin A. (August 2002). "Rare, structurally homologous self-peptides promote thymocyte positive selection". Immunity. 17 (2): 131–142. doi:10.1016/S1074-7613(02)00361-8. ISSN 1074-7613. PMID 12196285.
- 1 2 Sasaki, Katsuhiro; Takada, Kensuke; Ohte, Yuki; Kondo, Hiroyuki; Sorimachi, Hiroyuki; Tanaka, Keiji; Takahama, Yousuke; Murata, Shigeo (2015-06-23). "Thymoproteasomes produce unique peptide motifs for positive selection of CD8+ T cells". Nature Communications. 6 (1): 7484. Bibcode:2015NatCo...6.7484S. doi:10.1038/ncomms8484. ISSN 2041-1723. PMC 4557289. PMID 26099460.
- 1 2 Murata, Shigeo; Sasaki, Katsuhiro; Kishimoto, Toshihiko; Niwa, Shin-ichiro; Hayashi, Hidemi; Takahama, Yousuke; Tanaka, Keiji (2007-06-01). "Regulation of CD8+ T Cell Development by Thymus-Specific Proteasomes". Science. 316 (5829): 1349–1353. Bibcode:2007Sci...316.1349M. doi:10.1126/science.1141915. ISSN 0036-8075. PMID 17540904. S2CID 37185716.
- ↑ Nitta, Takeshi; Murata, Shigeo; Sasaki, Katsuhiro; Fujii, Hideki; Ripen, Adiratna Mat; Ishimaru, Naozumi; Koyasu, Shigeo; Tanaka, Keiji; Takahama, Yousuke (January 2010). "Thymoproteasome Shapes Immunocompetent Repertoire of CD8+ T Cells". Immunity. 32 (1): 29–40. doi:10.1016/j.immuni.2009.10.009. ISSN 1074-7613. PMID 20045355.
- ↑ Takada, Kensuke; Van Laethem, Francois; Xing, Yan; Akane, Kazuyuki; Suzuki, Haruhiko; Murata, Shigeo; Tanaka, Keiji; Jameson, Stephen C; Singer, Alfred (2015-08-24). "TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8+ T cells". Nature Immunology. 16 (10): 1069–1076. doi:10.1038/ni.3237. ISSN 1529-2908. PMC 4810782. PMID 26301566.
- 1 2 Nakagawa, T.; Roth, W.; Wong, P.; Nelson, A.; Farr, A.; Deussing, J.; Villadangos, J. A.; Ploegh, H.; Peters, C. (1998-04-17). "Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus". Science. 280 (5362): 450–453. Bibcode:1998Sci...280..450N. doi:10.1126/science.280.5362.450. ISSN 0036-8075. PMID 9545226.
- 1 2 Honey, Karen; Nakagawa, Terry; Peters, Christoph; Rudensky, Alexander (2002-05-20). "Cathepsin L Regulates CD4+ T Cell Selection Independently of Its Effect on Invariant Chain". The Journal of Experimental Medicine. 195 (10): 1349–1358. doi:10.1084/jem.20011904. ISSN 0022-1007. PMC 2193748. PMID 12021314.
- ↑ Bowlus, Christopher L.; Ahn, Jung; Chu, Tom; Gruen, Jeffrey R. (September 1999). "Cloning of a Novel MHC-Encoded Serine Peptidase Highly Expressed by Cortical Epithelial Cells of the Thymus". Cellular Immunology. 196 (2): 80–86. doi:10.1006/cimm.1999.1543. ISSN 0008-8749. PMID 10527559.
- ↑ Gommeaux, Julien; Grégoire, Claude; Nguessan, Prudence; Richelme, Mireille; Malissen, Marie; Guerder, Sylvie; Malissen, Bernard; Carrier, Alice (April 2009). "Thymus-specific serine protease regulates positive selection of a subset of CD4+thymocytes". European Journal of Immunology. 39 (4): 956–964. doi:10.1002/eji.200839175. ISSN 0014-2980. PMID 19283781.
- ↑ Mizushima, Noboru; Yamamoto, Akitsugu; Matsui, Makoto; Yoshimori, Tamotsu; Ohsumi, Yoshinori (March 2004). "In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker". Molecular Biology of the Cell. 15 (3): 1101–1111. doi:10.1091/mbc.e03-09-0704. ISSN 1059-1524. PMC 363084. PMID 14699058.
- ↑ Feng, Yuchen; He, Ding; Yao, Zhiyuan; Klionsky, Daniel J (2013-12-24). "The machinery of macroautophagy". Cell Research. 24 (1): 24–41. doi:10.1038/cr.2013.168. ISSN 1001-0602. PMC 3879710. PMID 24366339.
- ↑ Nedjic, Jelena; Aichinger, Martin; Emmerich, Jan; Mizushima, Noboru; Klein, Ludger (2008-08-13). "Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance". Nature. 455 (7211): 396–400. Bibcode:2008Natur.455..396N. CiteSeerX 10.1.1.655.8545. doi:10.1038/nature07208. ISSN 0028-0836. PMID 18701890. S2CID 4390542.
- ↑ Gordon, Julie; Wilson, Valerie A; Blair, Natalie F; Sheridan, Julie; Farley, Alison; Wilson, Linda; Manley, Nancy R; Blackburn, C Clare (2004-04-18). "Functional evidence for a single endodermal origin for the thymic epithelium". Nature Immunology. 5 (5): 546–553. doi:10.1038/ni1064. ISSN 1529-2908. PMID 15098031. S2CID 10282524.
- ↑ Rossi, Simona W.; Jenkinson, William E.; Anderson, Graham; Jenkinson, Eric J. (June 2006). "Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium". Nature. 441 (7096): 988–991. Bibcode:2006Natur.441..988R. doi:10.1038/nature04813. ISSN 0028-0836. PMID 16791197. S2CID 2358330.
- ↑ Bleul, Conrad C.; Corbeaux, Tatiana; Reuter, Alexander; Fisch, Paul; Mönting, Jürgen Schulte; Boehm, Thomas (June 2006). "Formation of a functional thymus initiated by a postnatal epithelial progenitor cell". Nature. 441 (7096): 992–996. Bibcode:2006Natur.441..992B. doi:10.1038/nature04850. ISSN 0028-0836. PMID 16791198. S2CID 4396922.
- ↑ Baik, Song; Jenkinson, Eric J.; Lane, Peter J. L.; Anderson, Graham; Jenkinson, William E. (2013-02-11). "Generation of both cortical and Aire+medullary thymic epithelial compartments from CD205+progenitors". European Journal of Immunology. 43 (3): 589–594. doi:10.1002/eji.201243209. ISSN 0014-2980. PMC 3960635. PMID 23299414.
- ↑ Ohigashi, Izumi; Zuklys, Saulius; Sakata, Mie; Mayer, Carlos E.; Zhanybekova, Saule; Murata, Shigeo; Tanaka, Keiji; Holländer, Georg A.; Takahama, Yousuke (2013-06-11). "Aire-expressing thymic medullary epithelial cells originate from β5t-expressing progenitor cells". Proceedings of the National Academy of Sciences. 110 (24): 9885–9890. Bibcode:2013PNAS..110.9885O. doi:10.1073/pnas.1301799110. PMC 3683726. PMID 23720310.
- ↑ Ohigashi, Izumi; Zuklys, Saulius; Sakata, Mie; Mayer, Carlos E.; Hamazaki, Yoko; Minato, Nagahiro; Hollander, Georg A.; Takahama, Yousuke (November 2015). "Adult Thymic Medullary Epithelium Is Maintained and Regenerated by Lineage-Restricted Cells Rather Than Bipotent Progenitors". Cell Reports. 13 (7): 1432–1443. doi:10.1016/j.celrep.2015.10.012. ISSN 2211-1247. PMID 26549457.
- ↑ Mayer, Carlos E.; Žuklys, Saulius; Zhanybekova, Saule; Ohigashi, Izumi; Teh, Hong-Ying; Sansom, Stephen N.; Shikama-Dorn, Noriko; Hafen, Katrin; Macaulay, Iain C. (2016-01-18). "Dynamic spatio-temporal contribution of single β5t+ cortical epithelial precursors to the thymus medulla". European Journal of Immunology. 46 (4): 846–856. doi:10.1002/eji.201545995. ISSN 0014-2980. PMC 4832341. PMID 26694097.
- ↑ Meireles, Catarina; Ribeiro, Ana R.; Pinto, Rute D.; Leitão, Catarina; Rodrigues, Pedro M.; Alves, Nuno L. (2017-04-13). "Thymic crosstalk restrains the pool of cortical thymic epithelial cells with progenitor properties". European Journal of Immunology. 47 (6): 958–969. doi:10.1002/eji.201746922. hdl:10216/111812. ISSN 0014-2980. PMID 28318017.
- ↑ Brunk, Fabian; Michel, Chloé; Holland-Letz, Tim; Slynko, Alla; Kopp-Schneider, Annette; Kyewski, Bruno; Pinto, Sheena (2017-05-22). "Dissecting and modeling the emergent murine TEC compartment during ontogeny". European Journal of Immunology. 47 (7): 1153–1159. doi:10.1002/eji.201747006. ISSN 0014-2980. PMID 28439878.
- ↑ Ucar, Ahmet; Ucar, Olga; Klug, Paula; Matt, Sonja; Brunk, Fabian; Hofmann, Thomas G.; Kyewski, Bruno (August 2014). "Adult Thymus Contains FoxN1− Epithelial Stem Cells that Are Bipotent for Medullary and Cortical Thymic Epithelial Lineages". Immunity. 41 (2): 257–269. doi:10.1016/j.immuni.2014.07.005. ISSN 1074-7613. PMC 4148705. PMID 25148026.
- ↑ Ulyanchenko, Svetlana; O’Neill, Kathy E.; Medley, Tanya; Farley, Alison M.; Vaidya, Harsh J.; Cook, Alistair M.; Blair, Natalie F.; Blackburn, C. Clare (March 2016). "Identification of a Bipotent Epithelial Progenitor Population in the Adult Thymus". Cell Reports. 14 (12): 2819–2832. doi:10.1016/j.celrep.2016.02.080. ISSN 2211-1247. PMC 4819909. PMID 26997270.
- ↑ Hamazaki, Yoko; Fujita, Harumi; Kobayashi, Takashi; Choi, Yongwon; Scott, Hamish S; Matsumoto, Mitsuru; Minato, Nagahiro (2007-02-04). "Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin". Nature Immunology. 8 (3): 304–311. doi:10.1038/ni1438. ISSN 1529-2908. PMID 17277780. S2CID 7775843.
- ↑ Sekai, Miho; Hamazaki, Yoko; Minato, Nagahiro (November 2014). "Medullary Thymic Epithelial Stem Cells Maintain a Functional Thymus to Ensure Lifelong Central T Cell Tolerance". Immunity. 41 (5): 753–761. doi:10.1016/j.immuni.2014.10.011. ISSN 1074-7613. PMID 25464854.