Methanotroph
Methanotrophs (sometimes called methanophiles) are prokaryotes that metabolize methane as their source of carbon and to unlock the energy of oxygen,[1] nitrate, sulfate, or other oxidized species. They are bacteria or archaea, can grow aerobically or anaerobically, and require single-carbon compounds to survive.
Methanotrophs are especially common in or near environments where methane is produced, although some methanotrophs can oxidize atmospheric methane. Their habitats include wetlands, soils, marshes, rice paddies, landfills, aquatic systems (lakes, oceans, streams) and more. They are of special interest to researchers studying global warming, as they play a significant role in the global methane budget, by reducing the amount of methane emitted to the atmosphere.[2][3]
Methanotrophy is a special case of methylotrophy, using single-carbon compounds that are more reduced than carbon dioxide. Some methylotrophs, however, can also make use of multi-carbon compounds; this differentiates them from methanotrophs, which are usually fastidious methane and methanol oxidizers. The only facultative methanotrophs isolated to date are members of the genus Methylocella silvestris,[4][5] Methylocapsa aurea[6] and several Methylocystis strains.[7]
In functional terms, methanotrophs are referred to as methane-oxidizing bacteria. However, methane-oxidizing bacteria encompass other organisms that are not regarded as sole methanotrophs. For this reason, methane-oxidizing bacteria have been separated into subgroups: methane-assimilating bacteria (MAB) groups, the methanotrophs, and autotrophic ammonia-oxidizing bacteria (AAOB), which cooxidize methane.[3]
Classification
Methanotrophs can be either bacteria or archaea. Which methanotroph species is present is mainly determined by the availability of electron acceptors. Many types of methane oxidizing bacteria (MOB) are known. Differences in the method of formaldehyde fixation and membrane structure divide these bacterial methanotrophs into several groups. There are several subgroups among the methanotrophic archaea.
Aerobic
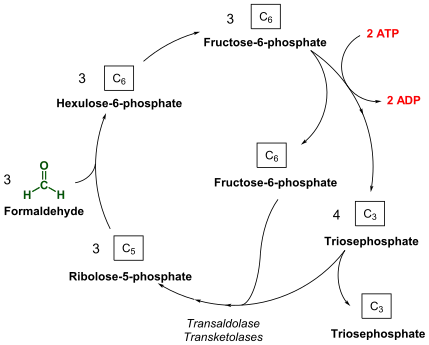
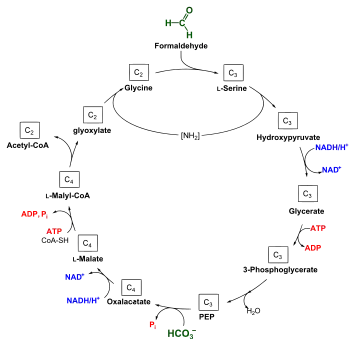
Under aerobic conditions, methanotrophs combine oxygen and methane to form formaldehyde, which is then incorporated into organic compounds via the serine pathway or the ribulose monophosphate (RuMP) pathway, and carbon dioxide, which is released. Type I and type X methanotrophs are part of the Gammaproteobacteria and they use the RuMP pathway to assimilate carbon. Type II methanotrophs are part of the Alphaproteobacteria and use the serine pathway of carbon assimilation. They also characteristically have a system of internal membranes within which methane oxidation occurs. Methanotrophs in Gammaproteobacteria are known from the family Methylococcaceae.[8] Methanotrophs from Alphaproteobacteria are found in families Methylocystaceae and Beijerinckiaceae.
Aerobic methanotrophs are also known from the Methylacidiphilaceae (phylum Verrucomicrobia).[9] In contrast to Gammaproteobacteria and Alphaproteobacteria, methanotrophs in the phylum Verrucomicrobia are mixotrophs.[10][11] In 2021 a bacterial bin from the phylum Gemmatimonadetes called Candidatus Methylotropicum kingii showing aerobic methanotrophy was discovered thus suggesting methanotrophy to be present in the four bacterial phyla.[12]
In some cases, aerobic methane oxidation can take place in anoxic environments. Candidatus Methylomirabilis oxyfera belongs to the phylum NC10 bacteria, and can catalyze nitrite reduction through an "intra-aerobic" pathway, in which internally produced oxygen is used to oxidise methane.[13][14] In clear water lakes, methanotrophs can live in the anoxic water column, but receive oxygen from photosynthetic organisms, which they then directly consume to oxidize methane.[15]
No aerobic methanotrophic archaea are known.
Anaerobic
Under anoxic conditions, methanotrophs use different electron acceptors for methane oxidation. This can happen in anoxic habitats such as marine or lake sediments, oxygen minimum zones, anoxic water columns, rice paddies and soils. Some specific methanotrophs can reduce nitrate,[16] nitrite,[17] iron,[18] sulfate,[19] or manganese ions and couple that to methane oxidation without syntrophic partner. Investigations in marine environments revealed that methane can be oxidized anaerobically by consortia of methane oxidizing archaea and sulfate-reducing bacteria.[20][21] This type of anaerobic oxidation of methane (AOM) mainly occurs in anoxic marine sediments. The exact mechanism is still a topic of debate but the most widely accepted theory is that the archaea use the reversed methanogenesis pathway to produce carbon dioxide and another, unknown intermediate, which is then used by the sulfate-reducing bacteria to gain energy from the reduction of sulfate to hydrogen sulfide and water.
The anaerobic methanotrophs are not related to the known aerobic methanotrophs; the closest cultured relatives to the anaerobic methanotrophs are the methanogens in the order Methanosarcinales.[22]
Special species
Methylococcus capsulatus is used to produce animal feed from natural gas.[23]
In 2010 a new bacterium Candidatus Methylomirabilis oxyfera from the phylum NC10 was identified that can couple the anaerobic oxidation of methane to nitrite reduction without the need for a syntrophic partner.[13] Based on studies of Ettwig et al.,[13] it is believed that M. oxyfera oxidizes methane anaerobically by utilizing oxygen produced internally from the dismutation of nitric oxide into nitrogen and oxygen gas.
Taxonomy
Many methanotrophic cultures have been isolated and formally characterized over the past 5 decades, starting with the classical study of Whittenbury (Whittenbury et al., 1970). Currently, 18 genera of cultivated aerobic methanotrophic Gammaproteobacteria and 5 genera of Alphaproteobacteria are known, represented by approx. 60 different species.[24]
Methane oxidation
Methanotrophs oxidize methane by first initiating reduction of an oxygen atom to H2O2 and transformation of methane to CH3OH using methane monooxygenases (MMOs).[25] Furthermore, two types of MMO have been isolated from methanotrophs: soluble methane monooxygenase (sMMO) and particulate methane monooxygenase (pMMO).
Cells containing pMMO have demonstrated higher growth capabilities and higher affinity for methane than sMMO containing cells.[25] It is suspected that copper ions may play a key role in both pMMO regulation and the enzyme catalysis, thus limiting pMMO cells to more copper-rich environments than sMMO producing cells.[26]
See also
- Borg (microbiology)
References
- ↑ Schmidt-Rohr, K. (2020). "Oxygen Is the High-Energy Molecule Powering Complex Multicellular Life: Fundamental Corrections to Traditional Bioenergetics". ACS Omega 5: 2221-2233. http://dx.doi.org/10.1021/acsomega.9b03352
- ↑ Oremland RS, Culbertson CW (1992). "Importance of methane-oxidizing bacteria in the methane budget as revealed by the use of a specific inhibitor". Nature. 356 (6368): 421–423. Bibcode:1992Natur.356..421O. doi:10.1038/356421a0. S2CID 4234351.
- 1 2 Holmes AJ, Roslev P, McDonald IR, Iversen N, Henriksen K, Murrell JC (August 1999). "Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake". Applied and Environmental Microbiology. 65 (8): 3312–8. Bibcode:1999ApEnM..65.3312H. doi:10.1128/AEM.65.8.3312-3318.1999. PMC 91497. PMID 10427012.
- ↑ Dedysh SN, Knief C, Dunfield PF (July 2005). "Methylocella species are facultatively methanotrophic". Journal of Bacteriology. 187 (13): 4665–70. doi:10.1128/JB.187.13.4665-4670.2005. PMC 1151763. PMID 15968078.
- ↑ Chen Y, Crombie A, Rahman MT, Dedysh SN, Liesack W, Stott MB, et al. (July 2010). "Complete genome sequence of the aerobic facultative methanotroph Methylocella silvestris BL2". Journal of Bacteriology. 192 (14): 3840–1. doi:10.1128/JB.00506-10. PMC 2897342. PMID 20472789.
- ↑ Dunfield PF, Belova SE, Vorob'ev AV, Cornish SL, Dedysh SN (November 2010). "Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa". International Journal of Systematic and Evolutionary Microbiology. 60 (Pt 11): 2659–2664. doi:10.1099/ijs.0.020149-0. PMID 20061505.
- ↑ Belova SE, Baani M, Suzina NE, Bodelier PL, Liesack W, Dedysh SN (February 2011). "Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp". Environmental Microbiology Reports. 3 (1): 36–46. doi:10.1111/j.1758-2229.2010.00180.x. PMID 23761229.
- ↑ Stein LY, Roy R, Dunfield PF (2012-04-16). "Aerobic Methanotrophy and Nitrification: Processes and Connections". eLS. Chichester, UK: John Wiley & Sons, Ltd. pp. a0022213. doi:10.1002/9780470015902.a0022213. ISBN 978-0-470-01617-6. Retrieved 2021-01-17.
- ↑ Op den Camp HJ, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, et al. (October 2009). "Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia". Environmental Microbiology Reports. 1 (5): 293–306. doi:10.1111/j.1758-2229.2009.00022.x. PMID 23765882.
- ↑ Carere CR, Hards K, Houghton KM, Power JF, McDonald B, Collet C, et al. (November 2017). "Mixotrophy drives niche expansion of verrucomicrobial methanotrophs". The ISME Journal. 11 (11): 2599–2610. doi:10.1038/ismej.2017.112. PMC 5649168. PMID 28777381.
- ↑ Sharp CE, Stott MB, Dunfield PF (2012). "Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing". Frontiers in Microbiology. 3: 303. doi:10.3389/fmicb.2012.00303. PMC 3421453. PMID 22912630.
- ↑ Bay SK, Dong X, Bradley JA, Leung PM, Grinter R, Jirapanjawat T, et al. (January 2021). "Trace gas oxidizers are widespread and active members of soil microbial communities". Nature Microbiology. 6 (2): 246–256. doi:10.1038/s41564-020-00811-w. PMID 33398096. S2CID 230663681.
- 1 2 3 Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, et al. (March 2010). "Nitrite-driven anaerobic methane oxidation by oxygenic bacteria" (PDF). Nature. 464 (7288): 543–8. Bibcode:2010Natur.464..543E. doi:10.1038/nature08883. PMID 20336137. S2CID 205220000.
- ↑ Zhu B, van Dijk G, Fritz C, Smolders AJ, Pol A, Jetten MS, Ettwig KF (December 2012). "Anaerobic oxidization of methane in a minerotrophic peatland: enrichment of nitrite-dependent methane-oxidizing bacteria". Applied and Environmental Microbiology. 78 (24): 8657–65. Bibcode:2012ApEnM..78.8657Z. doi:10.1128/AEM.02102-12. PMC 3502929. PMID 23042166.
- ↑ Milucka J, Kirf M, Lu L, Krupke A, Lam P, Littmann S, et al. (September 2015). "Methane oxidation coupled to oxygenic photosynthesis in anoxic waters". The ISME Journal. 9 (9): 1991–2002. doi:10.1038/ismej.2015.12. PMC 4542029. PMID 25679533.
- ↑ Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, et al. (August 2013). "Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage". Nature. 500 (7464): 567–70. Bibcode:2013Natur.500..567H. doi:10.1038/nature12375. PMID 23892779. S2CID 4368118.
- ↑ Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, et al. (March 2010). "Nitrite-driven anaerobic methane oxidation by oxygenic bacteria". Nature. 464 (7288): 543–8. Bibcode:2010Natur.464..543E. doi:10.1038/nature08883. PMID 20336137. S2CID 205220000.
- ↑ Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MS, Kartal B (November 2016). "Archaea catalyze iron-dependent anaerobic oxidation of methane". Proceedings of the National Academy of Sciences of the United States of America. 113 (45): 12792–12796. doi:10.1073/pnas.1609534113. PMC 5111651. PMID 27791118.
- ↑ Milucka J, Ferdelman TG, Polerecky L, Franzke D, Wegener G, Schmid M, et al. (November 2012). "Zero-valent sulphur is a key intermediate in marine methane oxidation". Nature. 491 (7425): 541–6. Bibcode:2012Natur.491..541M. doi:10.1038/nature11656. PMID 23135396. S2CID 32356495.
- ↑ Offre P, Spang A, Schleper C (2013-09-08). "Archaea in biogeochemical cycles". Annual Review of Microbiology. 67 (1): 437–57. doi:10.1146/annurev-micro-092412-155614. PMID 23808334.
- ↑ Thauer RK (June 2011). "Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2". Current Opinion in Microbiology. 14 (3): 292–9. doi:10.1016/j.mib.2011.03.003. PMID 21489863.
- ↑ Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. (October 2000). "A marine microbial consortium apparently mediating anaerobic oxidation of methane". Nature. 407 (6804): 623–6. Bibcode:2000Natur.407..623B. doi:10.1038/35036572. PMID 11034209. S2CID 205009562.
- ↑ Le Page M (2016-11-19). "Food made from natural gas will soon feed farm animals – and us". New Scientist. Retrieved 2016-12-11.
- ↑ Orata FD, Meier-Kolthoff JP, Sauvageau D, Stein LY (2018). "Phylogenomic Analysis of the Gammaproteobacterial Methanotrophs (Order Methylococcales) Calls for the Reclassification of Members at the Genus and Species Levels". Frontiers in Microbiology. 9: 3162. doi:10.3389/fmicb.2018.03162. PMC 6315193. PMID 30631317.
- 1 2 Hanson RS, Hanson TE (June 1996). "Methanotrophic bacteria". Microbiological Reviews. 60 (2): 439–71. doi:10.1128/MMBR.60.2.439-471.1996. PMC 239451. PMID 8801441.
- ↑ Lieberman RL, Rosenzweig AC (2004). "Biological methane oxidation: regulation, biochemistry, and active site structure of particulate methane monooxygenase". Critical Reviews in Biochemistry and Molecular Biology. 39 (3): 147–64. doi:10.1080/10409230490475507. PMID 15596549. S2CID 21628195.
External links
- Anaerobic oxidation of methane
- Methane-Eating Bug Holds Promise For Cutting Greenhouse Gas. Media Release, GNS Science, New Zealand