Mycobacteriophage

A mycobacteriophage is a member of a group of bacteriophages known to have mycobacteria as host bacterial species. While originally isolated from the bacterial species Mycobacterium smegmatis and Mycobacterium tuberculosis,[2] the causative agent of tuberculosis, more than 4,200 mycobacteriophage have since been isolated from various environmental and clinical sources. 2,042 have been completely sequenced.[3] Mycobacteriophages have served as examples of viral lysogeny and of the divergent morphology and genetic arrangement characteristic of many phage types.[4]
All mycobacteriophages found thus far have had double-stranded DNA genomes and have been classified by their structure and appearance into siphoviridae or myoviridae.[5]
Discovery
A bacteriophage found to infect Mycobacterium smegmatis in 1947 was the first documented example of a mycobacteriophage. It was found in cultures of the bacteria originally growing in moist compost.[6] The first bacteriophage that infects M. tuberculosis was discovered in 1954.[7]
Diversity
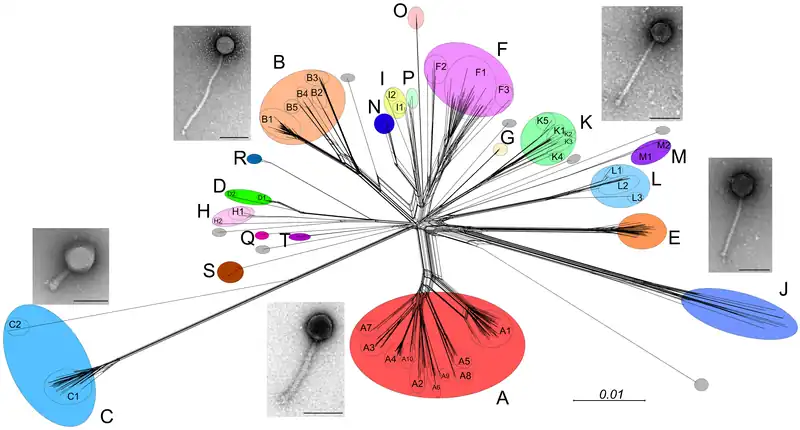
Thousands of mycobacteriophage have been isolated using a single host strain, Mycobacterium smegmatis mc2155, over 1400 of which have been completely sequenced.[3] These are mostly from environmental samples, but mycobacteriophages have also been isolated from stool samples of tuberculosis patients,[8] although these have yet to be sequenced.[9] About 30 distinct types (called clusters, or singletons if they have no relatives) that share little nucleotide sequence similarity have been identified. Many of the clusters span sufficient diversity that the genomes warrant division into subclusters (Figure 1).[9]
There is also considerable range in overall guanine plus cytosine content (GC%), from 50.3% to 70%, with an average of 64% (M. smegmatis is 67.3%). Thus, phage GC% does not necessarily match that of its host, and the consequent mismatch of codon usage profiles does not appear to be detrimental. Because new mycobacteriophages lacking extensive DNA similarity with the extant collection are still being discovered, and as there are at least seven singletons for which no relatives have been isolated, we clearly have yet to saturate the diversity of this particular population.[9]
The collection of >50,000 genes can be sorted into >3,900 groups (so-called phamilies, i.e. phage protein families) according to their shared amino acid sequences. Most of these phamilies (~75%) do not have homologues outside of the mycobacteriophages and are of unknown function. Genetic studies with mycobacteriophage Giles show that 45% of the genes are nonessential for lytic growth.[10]
Host range
Host range analysis shows that not all mycobacteriophages from M. smegmatis infect other strains and only phages in Cluster K and in certain subclusters of Cluster A efficiently infect M. tuberculosis (Figure 1).[11] However, mutants can be readily isolated from some phages that expand their host range to infect these other strains.[11] However, the molecular basis of host range depends on the behavior and presence of specific genes. This raises the probability of a correlation between gene phamilies and the preferred host.[11]
The realms of mycobacteriophage infection are not understood in its entirety because it involves various mechanisms including receptor availability, restriction-modification, abortive infection, and more. These mechanisms can be mediated through several processes like Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPRs) and the translational apparatus being modified. Phages overcome these constraints by evolving, spontaneous mutation, and diversifying.[11]
Genome architecture
The first sequenced mycobacteriophage genome was that of mycobacteriophage L5 in 1993.[12] In the following years hundreds of additional genomes have been sequenced.[3] Mycobacteriophages have highly mosaic genomes. Their genome sequences show evidence of extensive horizontal genetic transfer, both between phages and between phages and their mycobacterial hosts. Comparisons of these sequences have helped to explain how frequently genetic exchanges of this type may occur in nature, as well as how phages may contribute to bacterial pathogenicity.[13]
A selection of 60 mycobacteriophages were isolated and had their genomes sequenced in 2009. These genome sequences were grouped into clusters by several methods in an effort to determine similarities between the phages and to explore their genetic diversity. More than half of the phage species were originally found in or near Pittsburgh, Pennsylvania, though others were found in other United States locations, India, and Japan. No distinct differences were found in the genomes of mycobacteriophage species from different global origins.[14]
Mycobacteriophage genomes have been found to contain a subset of genes undergoing more rapid genetic flux than other elements of the genomes. These "rapid flux" genes are exchanged between mycobacteriophage more often and are 50 percent shorter in sequence than the average mycobacteriophage gene.[14]
Applications
Historically, mycobacteriophage have been used to "type" (i.e. "diagnose") mycobacteria, as each phage infects only one or a few bacterial strains.[15] In the 1980s phages were discovered as tools to genetically manipulate their hosts.[16] For instance, phage TM4 was used to construct shuttle phasmids that replicate as large cosmids in Escherichia coli and as phages in mycobacteria.[17] Shuttle phasmids can be manipulated in E. coli and used to efficiently introduce foreign DNA into mycobacteria.
Phages with mycobacterial hosts may be especially useful for understanding and fighting mycobacterial infections in humans. A system has been developed to use mycobacteriophage carrying a reporter gene to screen strains of M. tuberculosis for antibiotic resistance.[18] In the future, mycobacteriophage could be used to treat infections by phage therapy.[19][20]
References
- ↑ Padilla-Sanchez, Victor (24 July 2021), Mycobacteriophage ZoeJ Structural Model at Atomic Resolution, doi:10.5281/zenodo.5132914, retrieved 24 July 2021
- ↑ MANKIEWICZ, E. (1961). "Mycobacteriophages isolated from Persons with Tuberculous and Non-tuberculous Conditions". Nature. 191 (4796): 1416–1417. Bibcode:1961Natur.191.1416M. doi:10.1038/1911416b0. PMID 14469307. S2CID 4263411.
- 1 2 3 "Mycobacteriophage Database". Phagesdb.org. Retrieved 4 September 2017.
- ↑ Hatfull, Graham F. (13 October 2010). "Mycobacteriophages: Genes and Genomes". Annual Review of Microbiology. 64 (1): 331–356. doi:10.1146/annurev.micro.112408.134233. PMID 20528690.
- ↑ Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, et al. (January 2011). "Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution". PLOS ONE. 6 (1): e16329. Bibcode:2011PLoSO...616329P. doi:10.1371/journal.pone.0016329. PMC 3029335. PMID 21298013.
- ↑ Gardner, Grace M.; Weiser, Russell S. (1947). "A Bacteriophage for Mycobacterium smegmatis". Proceedings of the Society for Experimental Biology and Medicine. 66 (1): 205–206. doi:10.3181/00379727-66-16037. PMID 20270730. S2CID 20772051.
- ↑ Froman S, Will DW, Bogen E (October 1954). "Bacteriophage active against virulent Mycobacterium tuberculosis. I. Isolation and activity". Am J Public Health Nations Health. 44 (10): 1326–33. doi:10.2105/AJPH.44.10.1326. PMC 1620761. PMID 13197609.
- 1 2 3 4 Hatfull, G. F. (2014). "Mycobacteriophages: Windows into tuberculosis". PLOS Pathogens. 10 (3): e1003953. doi:10.1371/journal.ppat.1003953. PMC 3961340. PMID 24651299.
- ↑ Dedrick, R. M.; Marinelli, L. J.; Newton, G. L.; Pogliano, K; Pogliano, J; Hatfull, G. F. (2013). "Functional requirements for bacteriophage growth: Gene essentiality and expression in mycobacteriophage Giles". Molecular Microbiology. 88 (3): 577–89. doi:10.1111/mmi.12210. PMC 3641587. PMID 23560716.
- 1 2 3 4 Jacobs-Sera, D; Marinelli, L. J.; Bowman, C; Broussard, G. W.; Guerrero Bustamante, C; Boyle, M. M.; Petrova, Z. O.; Dedrick, R. M.; Pope, W. H.; Science Education Alliance Phage Hunters Advancing Genomics And Evolutionary Science Sea-Phages Program; Modlin, R. L.; Hendrix, R. W.; Hatfull, G. F. (2012). "On the nature of mycobacteriophage diversity and host preference". Virology. 434 (2): 187–201. doi:10.1016/j.virol.2012.09.026. PMC 3518647. PMID 23084079.
- ↑ Hatfull, G. F.; Sarkis, G. J. (1993). "DNA sequence, structure and gene expression of mycobacteriophage L5: A phage system for mycobacterial genetics". Molecular Microbiology. 7 (3): 395–405. doi:10.1111/j.1365-2958.1993.tb01131.x. PMID 8459766. S2CID 10188307.
- ↑ Pedulla, Marisa L; Ford, Michael E; Houtz, Jennifer M; Karthikeyan, Tharun; Wadsworth, Curtis; Lewis, John A; Jacobs-Sera; et al. (2003). "Origins of Highly Mosaic Mycobacteriophage Genomes". Cell. 113 (2): 171–182. doi:10.1016/S0092-8674(03)00233-2. PMID 12705866. S2CID 14055875.
- 1 2 Hatfull, Graham F.; Jacobs-Sera, Deborah; Lawrence, Jeffrey G.; Pope, Welkin H.; Russell, Daniel A.; Ko, Ching-Chung; Weber, Rebecca J.; Patel, Manisha C.; Germane, Katherine L.; Edgar, Robert H. (19 March 2010). "Comparative Genomic Analysis of 60 Mycobacteriophage Genomes: Genome Clustering, Gene Acquisition, and Gene Size". Journal of Molecular Biology. 397 (1): 119–143. doi:10.1016/j.jmb.2010.01.011. PMC 2830324. PMID 20064525.
- ↑ Jones Jr, W. D. (1975). "Phage typing report of 125 strains of "Mycobacterium tuberculosis"". Annali Sclavo; rivista di microbiologia e di immunologia. 17 (4): 599–604. PMID 820285.
- ↑ Jacobs WR Jr. 2000. Mycobacterium tuberculosis: a once genetically intractable organism. In Molecular Genetics of the Mycobacteria, ed. GF Hatfull, WR Jacobs Jr, pp. 1–16. Washington, DC: ASM Press
- ↑ Jacobs, W. R.; Tuckman, M.; Bloom, B. R. (1987). "Introduction of foreign DNA into mycobacteria using a shuttle phasmid". Nature. 327 (6122): 532–535. Bibcode:1987Natur.327..532J. doi:10.1038/327532a0. PMID 3473289. S2CID 9192407.
- ↑ Mulvey, MC; Sacksteder, KA; Einck, L; Nacy, CA (2012). "Generation of a Novel Nucleic Acid-Based Reporter System To Detect Phenotypic Susceptibility to Antibiotics in Mycobacterium tuberculosis". mBio. 3 (2): e00312–11. doi:10.1128/mBio.00312-11. PMC 3312217. PMID 22415006.
- ↑ Danelishvili L, Young LS, Bermudez LE (2006). "In vivo efficacy of phage therapy for Mycobacterium avium infection as delivered by a nonvirulent mycobacterium". Microb. Drug Resist. 12 (1): 1–6. doi:10.1089/mdr.2006.12.1. PMID 16584300.
- ↑ McNerney R, Traoré H (2005). "Mycobacteriophage and their application to disease control". J. Appl. Microbiol. 99 (2): 223–33. doi:10.1111/j.1365-2672.2005.02596.x. PMID 16033452. S2CID 43134099.