Nogalamycin
 | |
| Identifiers | |
|---|---|
CAS Number |
|
3D model (JSmol) |
|
| ChEBI | |
| ChemSpider | |
| ECHA InfoCard | 100.162.283 |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
InChI
| |
SMILES
| |
| Properties | |
Chemical formula |
C39H49NO16 |
| Molar mass | 787.80 g/mol |
| Hazards | |
| Occupational safety and health (OHS/OSH): | |
Main hazards |
cardiotoxic |
| Related compounds | |
Related compounds |
menogaril |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Nogalamycin is an anthracycline antibiotic produced by the soil bacteria Streptomyces nogalater. It has antitumor properties but it is also highly cardiotoxic. The less cardiotoxic semisynthetic analog menogaril was developed in the 1970s. Currently nogalamycin and menogaril are not used clinically.[1]
Biosynthesis
Anthracycline biosynthesis involves the construction of an aglycone core (by a type II polyketide synthase) to which one or more sugar residues are attached. Nogalamycin consists of three components:
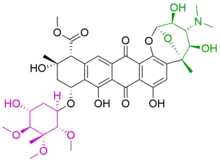
- nogalamycinone (black), the aglycone core
- nogalose (magenta)
- nogalamine (green)
Each component is built separately and then ligated together by a two glycosyltransferases.[2] All of the machinery associated with the biosynthesis of nogalamycin are located within the same biosynthetic gene cluster of S. nogalater.
Nogalamycinone biosynthesis
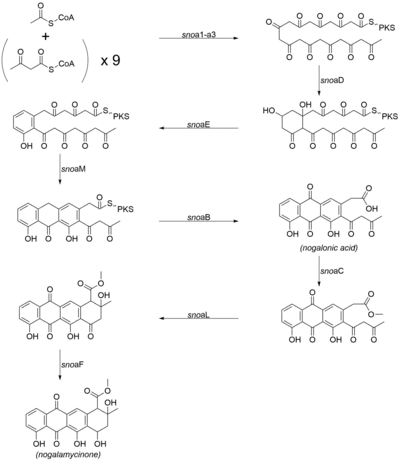
The biosynthetic pathway towards the aglycone core of nogalamycin has been determined by a combination of bioinformatic analysis and cloning of individual components of the biosynthetic pathway.[3][4][5] The biosynthetic route is similar to that of aklavinone (the aglycone core of most anthracyclines, including doxorubicin), the sole difference being that the first acyl group that is loaded into the PKS is an acetate rather than a propionate. The following genes are involved in the biosynthesis of the core nogalamycinone species:[3][6]
- snoa1 (ketosynthase-α)
- snoa2 (ketosynthase-β chain length factor)
- snoa3 (acyl carrier protein)
- snoaD (ketoreductase)
- snoaE (aromatase)
- snoaM (cyclase)
- snoaB (oxygenase)
- snoaC (methyltransferase)
- snoaL (cyclase)
- snoaF (ketoreductase)
Nogalamine and nogalose biosynthesis
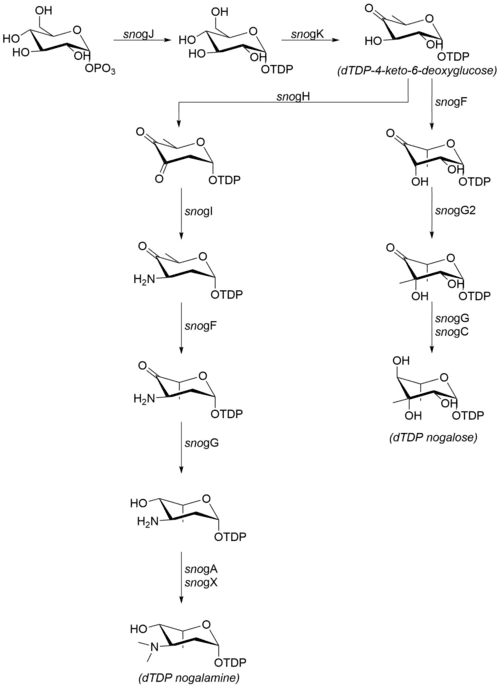
The sugar moieties that are attached to nogalamycinone are produced from glucose-1-phosphate. Although the steps following dTDP-4-keto-6-deoxyglucose have not been confirmed in vitro, the high degree of sequence similarity with homologous enzymes from other organisms suggests that the mechanism proceeds as detailed at right.[7] The following enzymes are involved in the biosynthesis of nogalamine and nogalose:[6]
- snogJ (dTDP-glucose synthase)
- snogK (4,6-dehydratase)
- snogF (3,5-epimerase)
- snogH (2,3-dehydratase)
- snogN (unknown)
- snogI (aminotransferase)
- snogG (ketoreductase)
- snogC (ketoreductase)
- snogA (N-methyltransferase)
- snogX (N-methyltransferase)
- snogG2 (C-methyltransferase)
Note that while much of the literature refers to the final, permethylated carbohydrate moiety as "nogalose",[8] more recent data suggest that the nogalose moiety on nogalamycin is methylated after the nogalamycinone core has been glycosylated.[2]
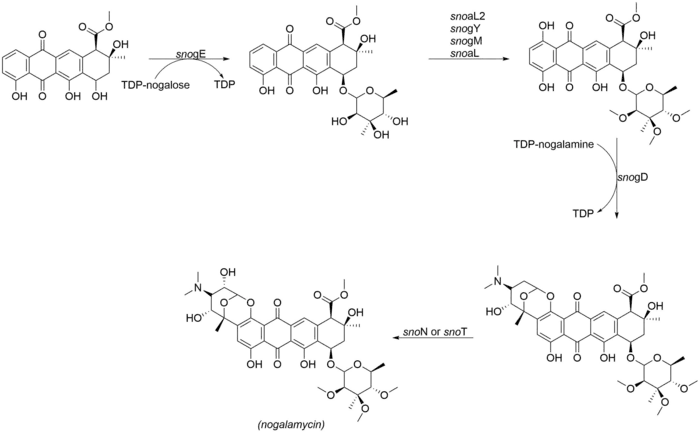
Nogalamycinone glycosylation and tailoring
The most noteworthy aspect of the structure of nogalamycin is the dual attachment of nogalamine both through O-glycosylation and also through a carbon-carbon bond at the C2 position of the nogalamycinone core,[2] The following enzymes are involved in the final tailoring steps of nogalamycin. SnoN and snoT are genes in the nogalamycin gene cluster that are likely to catalyze the final hydroxylation:
- snogE (glycosyltransferase)
- snoaL2 (hydroxylase)
- snogY (O-methyltransferase)
- snogM (putative O-methyltransferase)
- snogL (putative O-methyltransferase)
- snogD (glycosyltransferase)
- snoN/T
References
- ↑ NOGALAMYCIN. ChemicalBook.com. Accessed November 28, 2012.
- 1 2 3 Siitonen, V. et al. Identification of Late-Stage Glycosylation Steps in the Biosynthetic Pathway of the Anthracycline Nogalamycin. ChemBioChem 13, 120–128 (2011).
- 1 2 Torkkell, S. et al. The entire nogalamycin biosynthetic gene cluster of Streptomyces nogalater: characterization of a 20-kb DNA region and generation of hybrid structures. Mol Gen Genomics 266, 276–288 (2001).
- ↑ Räty, K. et al. Cloning and characterization of Streptomyces galilaeus aclacinomycins polyketide synthase (PKS) cluster. Gene 293, 115–122 (2002).
- ↑ Metsä-Ketelä, M., Palmu, K., Kunnari, T., Ylihonko, K. & Mäntsälä, P. Engineering Anthracycline Biosynthesis toward Angucyclines. Antimicrob. Agents Chemother. 47, 1291–1296 (2003).
- 1 2 Sultana, A. Mechanistic insights into the biosynthesis of polyketide antibiotics. (2006).
- ↑ K. Kharel, M., Lian, H. & Rohr, J. Characterization of the TDP-d-ravidosamine biosynthetic pathway: one-pot enzymatic synthesis of TDP-d-ravidosamine from thymidine-5-phosphate and glucose-1-phosphate. Organic & Biomolecular Chemistry 9, 1799–1808 (2011).
- ↑ Duchamp, D. J., Wiley, P. F., Hsiung, V. & Chidester, C. G. Structure, absolute configuration, and chemistry of nogalose. J. Org. Chem. 36, 2670–2673 (1971).