Pyoverdine
 | |
| Names | |
|---|---|
| Other names
Pyoverdin | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI |
|
PubChem CID |
|
InChI
| |
SMILES
| |
| Properties | |
Chemical formula |
C56H88N18O22 |
| Molar mass | 1365.424 g·mol−1 |
| Appearance | Solid |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Pyoverdines[1] (alternatively, and less commonly, spelled as pyoverdins) are fluorescent siderophores produced by certain pseudomonads.[2][3] Pyoverdines are important virulence factors, and are required for pathogenesis in many biological models of infection. Their contributions to bacterial pathogenesis include providing a crucial nutrient (i.e., iron), regulation of other virulence factors (including exotoxin A and the protease PrpL),[4] supporting the formation of biofilms,[5] and are increasingly recognized for having toxicity themselves.[6][7][8]
Pyoverdines have also been investigated as "Trojan Horse" molecules for the delivery of antimicrobials to otherwise resistant bacterial strains, as chelators that can be used for bioremediation of heavy metals, and as fluorescent reporters used to assay for the presence of iron and potentially other metals.[9]
Due to their bridging the gaps between pathogenicity, iron metabolism, and fluorescence, pyoverdines have piqued the curiosity of scientists around the world for over 100 years.
Biological functions
Like most siderophores, pyoverdine is synthesized and secreted into the environment when the microorganism that produces it detects that intracellular iron concentrations have fallen below a preset threshold. Although iron is the fourth-most abundant element in the Earth's crust, solubility of biologically relevant iron compounds is exceedingly low, and is generally insufficient for the needs of most (but not all) microorganisms. Siderophores, which are typically quite soluble and have exceptionally high avidity for iron (III) (the avidity of some siderophores for iron exceeds 1040 M-1 and many of the strongest avidities ever observed in nature are exhibited by siderophores for iron), help increase bioavailability of iron by pulling it into aqueous solution.
In addition to this role, pyoverdine has a number of other functions, including regulating virulence,[4][5] limiting the growth of other bacterial species (and serving as a sort of antimicrobial) by limiting iron availability, and sequestering other metals and preventing their toxicity.
Structure and characteristics
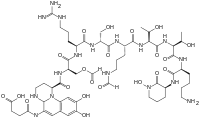
Although many (>100) forms of pyoverdine have been isolated and studied, they all have certain characteristics in common. Each pyoverdine molecule has three parts: a dihydroxyquinoline core, a 6-14 amino acid peptide that varies among strains, and a side chain (usually composed of a 4-5 carbon α-ketoacid from the Krebs/citric acid cycle). The core of pyoverdine is responsible for several of its properties, including its well-known yellowish color and fluorescence.
Structure
The dihydroxyquinoline core is composed of (1S)-5-amino-2,3-dihydro- 8,9-dihydroxy-1H-pyrimido[1,2-a]quinoline-1-carboxylic acid. This portion of the molecule is invariant amongst all observed pyoverdine molecules.
The core is modified by the addition of an amino acid chain of pyoverdine is composed of 6-14 amino acids. The chain of amino acids is built onto the chromophore core, and is synthesized via non-ribosomal peptide synthesis.[10][11] As is common for non-ribsosomally synthesized peptides, pyoverdine frequently includes D-form amino acids and non-standard amino acids, such as N-5-formyl-N-5-hydroxyornithine. The peptide chain may also be partially (or completely) cyclized. This peptide chain provides the other four aspects of the hexadentate interaction, usually through hydroxamate and/or hydroxycarboxylate groups. This portion of the molecule is also crucial for interaction with the ferripyoverdine receptor (FpvA) that allows ferripyoverdine to be imported into the cell. The peptide chain produced by a given strain of Pseudomonas is currently thought to be invariant.
Little is known about the particular function or importance of the ketoacid side chain, but it is well known[12] that pyoverdine molecules with different ketoacids (congeners) co-exist. Ketoacids that have been observed include succinate/succinamide, glutamate, glutarate, malate/malamide, and α-ketoglutarate.
| Pseudomonad species | Strain | Structure of the pyoverdine peptide chain |
|---|---|---|
| P. aeruginosa | ATCC15692 (PAO1) | Q-DSer-Arg-DSer-FoOHOrn-c(Lys-FoOHOrn-Thr-Thr) |
| P. aeruginosa | ATCC27853 | Q-DSer-FoOHDOrn-Orn-Gly-aDThr-Ser-cOHOrn |
| P. aeruginosa | Pa6 | Q-DSer-Dab-FoOHOrn-Gln-DGln-FoOHDOrn-Gly |
| P. chlororaphis | ATCC9446 | Q-DSer-Lys-Gly-FoOHOrn-c(Lys-FoOHDOrn-Ser) |
| P. fluorescens bv.I | ATCC13525 | Q-DSer-Lys-Gly-FoOHOrn-c(Lys-FoOHDOrn-Ser) |
| P. fluorescens bv.I | 9AW | Q-DSer-Lys-OHHis-aDThr-Ser-cOHOrn |
| P. fluorescens bv.III | ATCC17400 | Q-DAla-DLys-Gly-Gly-OHAsp-DGln/Dab-Ser-DAla-cOHOrn |
| P. fluorescens bv.V | 51W | Q-DAla-DLys-Gly-Gly-OHDAsp-DGln-DSer-Ala-Gly-aDThr-cOHOrn |
| P. fluorescens bv.V | 1W | Q-DSer-Lys-Gly-FoOHOrn-c(Lys-FoOHDOrn-Ser) |
| P. fluorescens bv.V | 10CW | Q-DSer-Lys-Gly-FoOHOrn-c(Lys-FoOHDOrn-Ser) |
| P. fluorescens bv.VI | PL7 | Q-DSer-AcOHDOrn-Ala-Gly-aDThr-Ala-cOHOrn |
| P. fluorescens bv.VI | PL8 | Q-DLys-AcOHDOrn-Ala-Gly-aDThr-Ser-cOHOrn |
| P. fluorescens | 1.3 | Q-DAla-DLys-Gly-Gly-OHAsp-DGln/Dab-Gly-Ser-cOHOrn |
| P. fluorescens | 18.1 | Q-DSer-Lys-Gly-FoOHOrn-Ser-DSer-Gly-c(Lys-FoOHDOrn-Ser) |
| P. fluorescens | CCM 2798 | Q-Ser-Dab-Gly-Ser-OHDAsp-Ala-Gly-DAla-Gly-cOHOrn |
| P. fluorescens | CFBP 2392 | Q-DLys-AcOHDOrn-Gly-aDThr-Thr-Gln-Gly-DSer-cOHOrn |
| P. fluorescens | CHA0 | Q-Asp-FoOHDOrn-Lys-c(Thr-Ala-Ala-FoOHDOrn-Lys) |
| P. putida bv. B | 9BW | Q-DSer-Lys-OHHis-aDThr-Ser-cOHOrn |
| P. putida | CFBP 2461 | Q-Asp-Lys-OHDAsp-Ser-aDThr-Ala-Thr-DLys-cOHOrn |
| P. tolaasii | NCPPB 2192 | Q-DSer-Lys-Ser-DSer-Thr-Ser-AcOHOrn-Thr-DSer-cOHDOrn |
Characteristics
Amongst their other notable characteristics, pyoverdines exhibit bright, relatively photostable fluorescence with characteristic excitation and emission spectra that are rapidly and strongly quenched upon binding their natural ligand, iron. Excitation and molar absorptivity show moderate pH dependence, but fluorescence is generally unaffected by pH variations. Unlike fluorescence, spectroscopic absorption shows little quenching upon iron-binding, suggesting that the mechanism for molecular relaxation is vibrational, rather than via electromagnetic radiation.
Pyoverdine coordinates a hexadentate (i.e., six-part) chelation of iron that involves six different oxygen atoms (2 from the dihyodroxyquinoline core and 2 from each of 2 different amino acids in the backbone). This results in a very tightly coordinated octahedral complex that efficiently prevents the ingress of water or other materials that may disrupt binding. Typically, ferric iron is removed from pyoverdine by reduction to the ferrous state, for which pyoverdine has a much lower (i.e., 109 M-1) avidity. This allows for the non-destructive removal of iron from pyoverdine. After reduction, the iron is "handed off" to other carriers that have increased affinity for ferrous iron, while the apopyoverdine is re-exported for continued use.
Pyoverdine is structurally similar to azobactin, from Azotobacter vinelandii, except that the latter possesses an extra urea ring.[13]
Synthesis
Biosynthesis
In Pseudomonas aeruginosa PAO1 there are 14 pvd genes involved in the biosynthesis of pyoverdine.[14]
Pyoverdine biosynthesis seems to be largely regulated through the activity of the alternate sigma factor PvdS which, in turn, is regulated both by the Fur system and by the intracellular sequestration of PvdS at the plasma membrane and away from the nucleoid by the repressor FpvI.
Despite significant investigation, relatively little is known about the biosynthesis of pyoverdine. For example, It remains unclear whether the biosynthesis of pyoverdine takes place as individual components (i.e., the core, the peptide chain, and the ketoacid) or if the core and the other parts are condensed as a beginning molecule (possibly by the PvdL protein) and then modified by other enzymes afterward. For reasons that remain unclear, pyoverdine biosynthesis is strongly inhibited by the anti-cancer therapeutic fluorouracil,[15] particularly through its ability to disrupt RNA metabolism.[16] Although production of pyoverdines varies from strain to strain, fluorescent Pseudomonas species have been shown to produce between 200 and 500 mg/L when grown in iron-depleted conditions.[17][18]
Core
There is some dispute about the origin of the fluorescent chromophore core. Originally, it was widely thought to be synthesized by the pvcABCD operon, as deletion of portions of the pvcC and pvcD genes disrupts pyoverdine production.[19] Like other aspects of pyoverdine biosynthesis, the regulation of the pvcABCD is iron-dependent, and the loss of these genes' activity resulted in pyoverdine disruption.
A separate report suggests that pvcABCD may be responsible for the synthesis of paerucumarin (a pseudoverdine-related molecule) instead, and claims that loss of activity in the locus has no effect on pyoverdine production.[20] In addition, some fluorescent Pseudomonads lack apparent homologs of these genes, further calling into question whether this is the function of these genes.
This is consistent with reports that pvdL combines coenzyme A to a myristic acid moiety, then adds a glutamate, D-tyrosine, and L-2,4-diaminobutyric acid (DAB).[21] An alternate biosynthetic pathway suggests that pvdL incorporates glutamate, 2,4,5-trihydroxyphenylalanine and L-2,4-daminobutyric acid instead.[22] This latter is supported by the identification of incorporation of a radiolabeled tyrosine into either pyoverdine or pseudoverdine.
This discrepancy remains unresolved.
Peptide chain
Several of the genes responsible for pyoverdine biosynthesis (e.g., pvdH, pvdA, and pvdF) are involved in the generation of precursor and alternate amino acids necessary for various portions of the molecule.[23] Several others (e.g., pvdI, and pvdJ) are directly responsible for "stitching" together the peptide chain.[23] pvdD terminates the chain and releases the precursor into the cytoplasm, which is consistent with identification of pyoverdine-like molecules in the cytoplasm with incompletely matured chromophores.[23]
Ketoacid
Currently, the best available evidence suggests that the ketoacid is originally attached to the chromophore core (as L-glutamate) when it is synthesized from D-tyrosine, L-2,4-diaminobutyric acid, and L-glutamate. It is unclear how this is later altered to the other congenerate (i.e., a-ketoglutarate, succinate/succinamide, etc.) forms.
Maturation and export
The localization of some of the Pvd proteins in the periplasm and the outer membrane (such as PvdN, PvdO, PvdP, and PvdQ) have been interpreted to suggest that portions of the maturation of pyoverdine takes place in this location, perhaps after being transported into the periplasm by PvdE, which is homologous to ABC type exporters. How completely matured pyoverdine is exported from the cell remains unclear. Once completely matured, pyoverdine is exported from the periplasm by PvdRT-OpmQ efflux pump.
Total chemical synthesis
A complete organic synthesis pathway for the pyoverdine produced by P. aeruginosa strain PAO1 has been reported[24] using solid-phase peptide synthesis. This protocol yielded pyoverdine at high yield (~48%) and is expected to substantially increase the ability of scientists to generate targeted derivatives on the pyoverdine scaffold and to facilitate the creation of siderophores with antimicrobial warheads.
Mechanisms of virulence
Pyoverdine has been reported to be required for virulence in a variety of disease models, including C. elegans and various models of murine infection (e.g., burn models, pneumonia models, etc.).[6][15][25]
As noted above, pyoverdine contributes in several fashions to general virulence, including regulating the production of itself, exotoxin A (which stalls translation), and the protease PrpL.[4] There is also evidence that, although not essential for its formation, pyoverdine contributes to the production and development of biofilms that are important for virulence.[5]
Finally, pyoverdine is associated with several types of toxicity in its own right. In 2001, Albesa and colleagues reported that pyoverdine purified from a strain of P. fluorescens exhibited profound cytotoxicity to mammalian macrophages and that this effect was at least partially dependent upon reactive oxygen species.[26] Later, Kirienko and colleagues determined that pyoverdine is both necessary and sufficient for killing C. elegans, that enters host cells, destabilizes mitochondrial dynamics, and induces a hypoxic response.[6][7] Exposure triggers a response that is consistent with hypoxia that depends on the HIF-1 protein, suggesting that the host perceives a condition where it lacks the molecular tools for generating ATP (generally, iron, oxygen, and cellular reducing equivalents).[6][7]
Role in microbial cooperation
Once pyoverdine is secreted, it diffuses freely in the environment. Iron-bound pyoverdine (also known as ferripyoverdine) can be taken up by any bacterial cell with the appropriate receptor, although this varies between strains.[27] Importantly, this creates a common good which can be exploited by 'cheaters' which retain the ability to use pyoverdine but have stopped making it. Since pyoverdine production is energetically costly, this can create a fitness advantage in cells that are not synthesizing it.[28][29][30][31] Consequently, pyoverdine has become a model trait to study microbial cooperation and exploitation.[32][33]
In P. aeruginosa, pyoverdine non-producing “cheat” bacteria have been shown to i) evolve readily from a producing ancestor;[34] and ii) outcompete cooperating strains in mixed culture in a density- and frequency-dependent manner.[35][36] Since pyoverdine usage relies on passive diffusion and pyoverdine production is metabolically costly, environmental conditions are known to influence the likelihood of successful exploitation. The competitive advantage of pyoverdine non-producers over producers in mixed culture was shown to be maximized when environments are well-mixed and molecules diffuse readily (low spatial structure) and when the costs and benefits of pyoverdine production are high, i.e. when iron is strongly limited.[30][37] Most studies on pyoverdine cooperation and cheating have been conducted using clinical isolates, but siderophore exploitation was recently also demonstrated in natural Pseudomonas isolates from non-clinical samples.[38][39]
Nomenclature
Currently, no widespread and systematic nomenclature is used to differentiate pyoverdine structures. A system was proposed in 1989,[40] consisting of Pyoverdine Type I, Type IIa, Type IIb, and Type III. At the time, only a few pyoverdine structures were known, and it was anticipated that much less variation would occur than has been seen. As a consequence of the tremendous heterogeneity observed in the peptide backbone, and the observation of congeners (pyoverdines from a single strain differing only in their ketoacid portions), nomenclature of pyoverdines remains rather tenuous and no single system has garnered universal acceptance.
History
- 1850s: Sédillot notes a blue-green discharge from surgical wound dressings.
- 1860: Pyoverdine (although not so named) was extracted from wound dressings by Fordos.
- 1862: Lucke associates pyoverdine with bacilli observed under microscope.
- 1882: Pseudomonas aeruginosa grown for first time in pure culture by Carle Gessard, reported in "On the Blue and Green Coloration of Bandages". Gessard names the organism Bacillus aeruginosa, after "aerugo", the Latin word for verdigris.
- 1889: Bouchard observes that injection of a rabbit infected with Bacillus anthracis (causative agent of anthrax) with P. aeruginosa prevents formation of anthrax.
- 1889: Bouchard discovers that pyoverdine fluoresces under ultraviolet light.
- 1948, 1952: First observations that concentrations of iron and pyoverdine are reciprocal.
- 1978: Meyer and colleagues make first demonstration of role for pyoverdine in iron acquisition.
- 1980s–1990s: First structures and regulation of proverdine worked out
- 1999: First determination that pyoverdine fluorescence is quenched by iron binding.
Other uses
Pseudoverdine
A compound related to pyoverdine, called pseudoverdine (formally known as 3-formylamino-6,7-dihydroxycoumarin) is also produced by some fluorescent Pseudomonads.[41] It is thought that pseudoverdine and pyoverdine may arise from a common precursor, 2,4,5-trihydroxyphenylalanine, which may condense with L-2,4-diaminobutyric acid to initiate pyoverdine production.[41]
Pseudoverdine is relatively similar to pyoverdine in its fluorescence and other spectroscopic properties, and its ability to chelate ferric iron, albeit at much lower affinity.[41] Unlike pyoverdine, it is incapable of transporting iron into cells, likely due to the absence of the peptide chain.[41] Another dissimilarity is that pseudoverdine does not appear to be regulated by the same processes as pyoverdine.[41]
References
- ↑ For the purposes of this page, pyoverdine will generally refer (unless otherwise noted) to the pyoverdine produced by Pseudomonas aeruginosa strain PAO1. It has been subjected to the most extensive study and can be considered the prototypical siderophore.
- ↑ S. Wendenbaum; P. Demange; A. Dell; J. M. Meyer; M. A. Abdallah (1983). "The structure of pyoverdine Pa, the siderophore of Pseudomonas aeruginosa". Tetrahedron Letters. 24 (44): 4877–4880. doi:10.1016/S0040-4039(00)94031-0.
- ↑ Menhart, N.; Thariath, A.; Viswanatha, T. (1991). "Characterization of the pyoverdines of Azotobacter vinelandii ATCC 12837 with regard to heterogeneity". Biology of Metals. 4 (4): 223–32. doi:10.1007/bf01141185. PMID 1838001. S2CID 8712926.
- 1 2 3 Lamont, Iain L.; Beare, Paul A.; Ochsner, Urs; Vasil, Adriana I.; Vasil, Michael L. (2002-05-14). "Siderophore-mediated signaling regulates virulence factor production in Pseudomonasaeruginosa". Proceedings of the National Academy of Sciences of the United States of America. 99 (10): 7072–7077. Bibcode:2002PNAS...99.7072L. doi:10.1073/pnas.092016999. ISSN 0027-8424. PMC 124530. PMID 11997446.
- 1 2 3 Banin, Ehud; Vasil, Michael L.; Greenberg, E. Peter (2005-08-02). "Iron and Pseudomonas aeruginosa biofilm formation". Proceedings of the National Academy of Sciences of the United States of America. 102 (31): 11076–11081. Bibcode:2005PNAS..10211076B. doi:10.1073/pnas.0504266102. ISSN 0027-8424. PMC 1182440. PMID 16043697.
- 1 2 3 4 Kirienko, Natalia V.; Kirienko, Daniel R.; Larkins-Ford, Jonah; Wählby, Carolina; Ruvkun, Gary; Ausubel, Frederick M. (2013-04-17). "Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death". Cell Host & Microbe. 13 (4): 406–416. doi:10.1016/j.chom.2013.03.003. ISSN 1934-6069. PMC 3641844. PMID 23601103.
- 1 2 3 Kirienko, Natalia V.; Ausubel, Frederick M.; Ruvkun, Gary (2015-02-10). "Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa". Proceedings of the National Academy of Sciences of the United States of America. 112 (6): 1821–1826. Bibcode:2015PNAS..112.1821K. doi:10.1073/pnas.1424954112. ISSN 1091-6490. PMC 4330731. PMID 25624506.
- ↑ Minandri, Fabrizia; Imperi, Francesco; Frangipani, Emanuela; Bonchi, Carlo; Visaggio, Daniela; Facchini, Marcella; Pasquali, Paolo; Bragonzi, Alessandra; Visca, Paolo (2016-08-01). "Role of Iron Uptake Systems in Pseudomonas aeruginosa Virulence and Airway Infection". Infection and Immunity. 84 (8): 2324–2335. doi:10.1128/IAI.00098-16. ISSN 1098-5522. PMC 4962624. PMID 27271740.
- ↑ Yoder, Michael F.; Kisaalita, William S. (2011-01-01). "Iron specificity of a biosensor based on fluorescent pyoverdin immobilized in sol-gel glass". Journal of Biological Engineering. 5: 4. doi:10.1186/1754-1611-5-4. ISSN 1754-1611. PMC 3114707. PMID 21554740.
- ↑ Hohlneicher, U.; Schäfer, M.; Fuchs, R.; Budzikiewicz, H. (2016-10-11). "Ferribactins as the biosynthetic precursors of the Pseudomonas siderophores pyoverdins". Zeitschrift für Naturforschung C. 56 (3–4): 308–310. doi:10.1515/znc-2001-3-423. ISSN 0939-5075. PMID 11371026.
- ↑ Visca, Paolo; Imperi, Francesco; Lamont, Iain L. (2007-01-01). "Pyoverdine siderophores: from biogenesis to biosignificance". Trends in Microbiology. 15 (1): 22–30. doi:10.1016/j.tim.2006.11.004. ISSN 0966-842X. PMID 17118662.
- ↑ Budzikiewicz, H (1997). "Siderophores of Fluorescent Pseudomonads". Zeitschrift für Naturforschung C. 52 (11–12): 713–720. doi:10.1515/znc-1997-11-1201. PMID 9463934. S2CID 26196277.
- 1 2 Meyer, J. M. (2000). "Pyoverdines: Pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species". Archives of Microbiology. 174 (3): 135–142. doi:10.1007/s002030000188. PMID 11041343. S2CID 13283224.
- ↑ Lamont, I. L.; Martin, L. W. (2003). "Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa". Microbiology. 149 (4): 833–842. doi:10.1099/mic.0.26085-0. PMID 12686626.
- 1 2 Imperi, Francesco; Massai, Francesco; Facchini, Marcella; Frangipani, Emanuela; Visaggio, Daniela; Leoni, Livia; Bragonzi, Alessandra; Visca, Paolo (2013-04-30). "Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity". Proceedings of the National Academy of Sciences of the United States of America. 110 (18): 7458–7463. Bibcode:2013PNAS..110.7458I. doi:10.1073/pnas.1222706110. ISSN 1091-6490. PMC 3645532. PMID 23569238.
- ↑ Kirienko, Daniel R.; Revtovich, Alexey V.; Kirienko, Natalia V. (2016-08-01). "A High-Content, Phenotypic Screen Identifies Fluorouridine as an Inhibitor of Pyoverdine Biosynthesis and Pseudomonas aeruginosa Virulence". mSphere. 1 (4): e00217–16. doi:10.1128/mSphere.00217-16. ISSN 2379-5042. PMC 4999921. PMID 27579370.
- ↑ Cornelis, P; Hohnadel, D; Meyer, JM (1989). "Evidence for different pyoverdine-mediated iron uptake systems among Pseudomonas aeruginosa strains". Infect Immun. 57 (11): 3491–3497. doi:10.1128/IAI.57.11.3491-3497.1989. PMC 259858. PMID 2509364.
- ↑ Hohnadel, Dany; Haas, Dieter; Meyer, Jean-Marie (1986-09-01). "Mapping of mutations affecting pyoverdine production in Pseudomonas aeruginosa". FEMS Microbiology Letters. 36 (2–3): 195–199. doi:10.1111/j.1574-6968.1986.tb01695.x. ISSN 0378-1097.
- ↑ Stintzi, A.; Johnson, Z.; Stonehouse, M.; Ochsner, U.; Meyer, J. M.; Vasil, M. L.; Poole, K. (1999-07-01). "The pvc gene cluster of Pseudomonas aeruginosa: role in synthesis of the pyoverdine chromophore and regulation by PtxR and PvdS". Journal of Bacteriology. 181 (13): 4118–4124. doi:10.1128/JB.181.13.4118-4124.1999. ISSN 0021-9193. PMC 93907. PMID 10383985.
- ↑ Clarke-Pearson, Michael F.; Brady, Sean F. (2008-10-01). "Paerucumarin, a new metabolite produced by the pvc gene cluster from Pseudomonas aeruginosa". Journal of Bacteriology. 190 (20): 6927–6930. doi:10.1128/JB.00801-08. ISSN 1098-5530. PMC 2566218. PMID 18689486.
- ↑ Budzikiewicz, H (1993). "Secondary metabolites from fluorescent pseudomonads". FEMS Microbiol Rev. 10 (3–4): 209–228. doi:10.1111/j.1574-6968.1993.tb05868.x. PMID 8318257.
- ↑ Stintzi, A; Cornelis, P; Hohnadel, D; Meyer, JM; Dean, C; Poole, K; Kourambas, S; Krishnapillai, V (1996). "Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa PAO". Microbiology. 142 (5): 1181–1190. doi:10.1099/13500872-142-5-1181. PMID 8704959.
- 1 2 3 Cézard, C.; Farvacques, N.; Sonnet, P. (2015-01-01). "Chemistry and biology of pyoverdines, Pseudomonas primary siderophores". Current Medicinal Chemistry. 22 (2): 165–186. doi:10.2174/0929867321666141011194624. ISSN 1875-533X. PMID 25312210.
- ↑ Mashiach, Roi; Meijler, Michael M. (2013-04-05). "Total synthesis of pyoverdin D". Organic Letters. 15 (7): 1702–1705. doi:10.1021/ol400490s. ISSN 1523-7052. PMID 23530778.
- ↑ Takase, H.; Nitanai, H.; Hoshino, K.; Otani, T. (2000-04-01). "Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice". Infection and Immunity. 68 (4): 1834–1839. doi:10.1128/iai.68.4.1834-1839.2000. ISSN 0019-9567. PMC 97355. PMID 10722571.
- ↑ Becerra, C.; Albesa, I.; Eraso, A. J. (2001-07-13). "Leukotoxicity of Pyoverdin, Production of Reactive Oxygen Species, and Effect of UV Radiation". Biochemical and Biophysical Research Communications. 285 (2): 414–418. doi:10.1006/bbrc.2001.5188. PMID 11444858.
- ↑ Bodilis, Josselin; Ghysels, Bart; Osayande, Julie; Matthijs, Sandra; Pirnay, Jean-Paul; Denayer, Sarah; De Vos, Daniel; Cornelis, Pierre (2009-08-01). "Distribution and evolution of ferripyoverdine receptors in Pseudomonas aeruginosa". Environmental Microbiology. 11 (8): 2123–2135. doi:10.1111/j.1462-2920.2009.01932.x. ISSN 1462-2920. PMID 19397675.
- ↑ Weigert, Michael; Kümmerli, Rolf (2017-07-12). "The physical boundaries of public goods cooperation between surface-attached bacterial cells". Proc. R. Soc. B. 284 (1858): 20170631. doi:10.1098/rspb.2017.0631. ISSN 0962-8452. PMC 5524494. PMID 28701557.
- ↑ Buckling, Angus; Harrison, Freya; Vos, Michiel; Brockhurst, Michael A.; Gardner, Andy; West, Stuart A.; Griffin, Ashleigh (2007-11-01). "Siderophore-mediated cooperation and virulence in Pseudomonas aeruginosa". FEMS Microbiology Ecology. 62 (2): 135–141. doi:10.1111/j.1574-6941.2007.00388.x. ISSN 0168-6496. PMID 17919300.
- 1 2 Kümmerli, Rolf; Brown, Sam P. (2010-11-02). "Molecular and regulatory properties of a public good shape the evolution of cooperation". Proceedings of the National Academy of Sciences. 107 (44): 18921–18926. Bibcode:2010PNAS..10718921K. doi:10.1073/pnas.1011154107. ISSN 0027-8424. PMC 2973908. PMID 20944065.
- ↑ Griffin, Ashleigh S.; West, Stuart A.; Buckling, Angus (2004). "Cooperation and competition in pathogenic bacteria". Nature. 430 (7003): 1024–1027. Bibcode:2004Natur.430.1024G. doi:10.1038/nature02744. hdl:1842/698. ISSN 1476-4687. PMID 15329720. S2CID 4429250.
- ↑ West, Stuart A.; Griffin, Ashleigh S.; Gardner, Andy; Diggle, Stephen P. (2006). "Social evolution theory for microorganisms". Nature Reviews Microbiology. 4 (8): 597–607. doi:10.1038/nrmicro1461. ISSN 1740-1534. PMID 16845430. S2CID 18451640.
- ↑ Kümmerli, R.; Santorelli, L. A.; Granato, E. T.; Dumas, Z.; Dobay, A.; Griffin, A. S.; West, S. A. (2015-12-01). "Co-evolutionary dynamics between public good producers and cheats in the bacterium Pseudomonas aeruginosa" (PDF). Journal of Evolutionary Biology. 28 (12): 2264–2274. doi:10.1111/jeb.12751. ISSN 1420-9101. PMID 26348785. S2CID 826683.
- ↑ Dumas, Z.; Kümmerli, R. (2012-03-01). "Cost of cooperation rules selection for cheats in bacterial metapopulations". Journal of Evolutionary Biology. 25 (3): 473–484. doi:10.1111/j.1420-9101.2011.02437.x. ISSN 1420-9101. PMID 22168669. S2CID 19132153.
- ↑ Ross‐Gillespie, Adin; Gardner, Andy; West, Stuart A.; Griffin, Ashleigh S. (2007-09-01). "Frequency Dependence and Cooperation: Theory and a Test with Bacteria". The American Naturalist. 170 (3): 331–342. doi:10.1086/519860. ISSN 0003-0147. PMID 17879185. S2CID 14248496.
- ↑ Ross-Gillespie, Adin; Gardner, Andy; Buckling, Angus; West, Stuart A.; Griffin, Ashleigh S. (2009-09-01). "Density Dependence and Cooperation: Theory and a Test with Bacteria". Evolution. 63 (9): 2315–2325. doi:10.1111/j.1558-5646.2009.00723.x. ISSN 1558-5646. PMID 19453724. S2CID 5699402.
- ↑ Kümmerli, Rolf; Griffin, Ashleigh S.; West, Stuart A.; Buckling, Angus; Harrison, Freya (2009-10-07). "Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa". Proceedings of the Royal Society of London B: Biological Sciences. 276 (1672): 3531–3538. doi:10.1098/rspb.2009.0861. ISSN 0962-8452. PMC 2817189. PMID 19605393.
- ↑ Bruce, John B.; Cooper, Guy A.; Chabas, Hélène; West, Stuart A.; Griffin, Ashleigh S. (2017-10-01). "Cheating and resistance to cheating in natural populations of the bacterium Pseudomonas fluorescens". Evolution. 71 (10): 2484–2495. doi:10.1111/evo.13328. ISSN 1558-5646. PMID 28833073. S2CID 3485902.
- ↑ Butaitė, Elena; Baumgartner, Michael; Wyder, Stefan; Kümmerli, Rolf (2017-09-04). "Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities". Nature Communications. 8 (1): 414. Bibcode:2017NatCo...8..414B. doi:10.1038/s41467-017-00509-4. ISSN 2041-1723. PMC 5583256. PMID 28871205.
- ↑ Briskot, G.; Taraz, K.; Budzikiewicz, H. (1989). "Bacterial Constituents, XXXVII. Pyoverdin-Type Siderophores from Pseudomonas aeruginosa". Liebigs Ann Chem. 1989 (4): 375–384. doi:10.1002/jlac.198919890164.
- 1 2 3 4 5 Longerich, I; Taraz, K; Budzikiewicz, H; Tsai, L; Meyer, JM (1993). "Pseudoverdin, a compound related to the pyoverdin chromophore from a Pseudomonas aeruginosa strain incapable to produce pyoverdins". Z Naturforsch C. 48 (5–6): 425–429. doi:10.1515/znc-1993-5-605. PMID 8363709. S2CID 29269780.