Reactive oxygen species
_%E2%80%93_some_examples_with_Lewis_structures.png.webp)
Reactive oxygen species (ROS) are highly reactive chemicals formed from O2. Examples of ROS include peroxides, superoxide, hydroxyl radical, singlet oxygen,[1] and alpha-oxygen.
The reduction of molecular oxygen (O2) produces superoxide (•O−
2), which is the precursor to most other reactive oxygen species:[2]
- O2 + e− → •O−
2
Dismutation of superoxide produces hydrogen peroxide (H2O2):[2]
- 2 H+ + •O−
2 + •O−
2 → H2O2 + O2
Hydrogen peroxide in turn may be partially reduced, thus forming hydroxide ions and hydroxyl radicals (•OH), or fully reduced to water:[2]
- H2O2 + e− → HO− + •OH
- 2 H+ + 2 e− + H2O2 → 2 H2O
In a biological context, ROS are byproducts of the normal metabolism of oxygen. ROS have roles in cell signaling and homeostasis.[3][4] ROS are intrinsic to cellular functioning, and are present at low and stationary levels in normal cells.[5] In vegetables, ROS are involved in metabolic processes related to photoprotection and tolerance to various types of stress.[6] However, ROS can cause irreversible damage to DNA as they oxidize and modify some cellular components and prevent them from performing their original functions. This suggests that ROS has a dual role; whether they will act as harmful, protective or signaling factors depends on the balance between ROS production and disposal at the right time and place.[7] In other words, oxygen toxicity can arise both from uncontrolled production and from the inefficient elimination of ROS by the antioxidant system. During times of environmental stress (e.g., UV or heat exposure), ROS levels can increase dramatically.[3] This may result in significant damage to cell structures. Cumulatively, this is known as oxidative stress. The production of ROS is strongly influenced by stress factor responses in plants, these factors that increase ROS production include drought, salinity, chilling, defense of pathogens, nutrient deficiency, metal toxicity and UV-B radiation. ROS are also generated by exogenous sources such as ionizing radiation[8] generating irreversible effects in the development of tissues in both animals and plants.[9]
Sources of ROS production
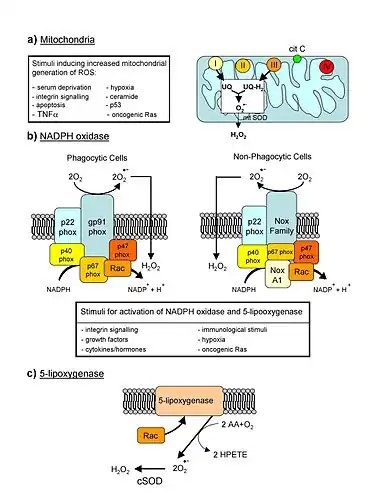
Endogenous sources
ROS are produced by biochemical reactions that occur during the processes of respiration and photosynthesis in organelles such as mitochondria, peroxisomes and chloroplasts.[7][12][13][14] During the respiration process the mitochondria convert energy for the cell into a usable form, adenosine triphosphate (ATP). The process of ATP production in the mitochondria, called oxidative phosphorylation, involves the transport of protons (hydrogen ions) across the inner mitochondrial membrane by means of the electron transport chain. In the electron transport chain, electrons are passed through a series of proteins via oxidation-reduction reactions, with each acceptor protein along the chain having a greater reduction potential than the previous. The last destination for an electron along this chain is an oxygen molecule. In normal conditions, the oxygen is reduced to produce water; however, in about 0.1–2% of electrons passing through the chain (this number derives from studies in isolated mitochondria, though the exact rate in live organisms is yet to be fully agreed upon), oxygen is instead prematurely and incompletely reduced to give the superoxide radical (•O−
2), most well documented for Complex I and Complex III.[15]
Another source of ROS production in animal cells is the electron transfer reactions catalyzed by the mitochondrial P450 systems in steroidogenic tissues.[16] These P450 systems are dependent on the transfer of electrons from NADPH to P450. During this process, some electrons "leak" and react with O2 producing superoxide. To cope with this natural source of ROS, the steroidogenic tissues, ovary and testis, have a large concentration of antioxidants such as vitamin C (ascorbate) and β-carotene and anti-oxidant enzymes.[17]
If too much damage is present in mitochondria, a cell undergoes apoptosis or programmed cell death.[18][19]
In addition, ROS are produced in immune cell signaling via the NOX pathway. Phagocytic cells such as neutrophils, eosinophils, and mononuclear phagocytes produce ROS when stimulated.[20][21]
In chloroplasts, the carboxylation and oxygenation reactions catalyzed by rubisco ensure that the functioning of the electron transport chain (ETC) occurs in an environment rich in O2. The leakage of electrons in the ETC will inevitably produce ROS within the chloroplasts.[7] ETC in photosystem I (PSI) was once believed to be the only source of ROS in chloroplasts. According to Huang et al.[22] the flow of electrons from the excited reaction centers is directed to the NADP and these are reduced to NADPH, and then they enter the Calvin cycle and reduce the final electron acceptor, CO2. In cases where there is an ETC overload, part of the electron flow is diverted from ferredoxin to O2, forming the superoxide free radical (by the Mehler reaction). In addition, electron leakage to O2 can also occur from the 2Fe-2S and 4Fe-4S clusters in the PSI ETC. However, PSII also provides electron leakage locations (QA, QB) for O2-producing O2-.[23][24] Superoxide (O2-) is generated from PSII, instead of PSI; QB is shown as the location for the generation of O2•-.[23]
Exogenous sources
The formation of ROS can be stimulated by a variety of agents such as pollutants, heavy metals,[11] tobacco, smoke, drugs, xenobiotics, or radiation. In plants, in addition to the action of dry abiotic factors, high temperature, interaction with other living beings can influence the production of ROS.
Ionizing radiation can generate damaging intermediates through the interaction with water, a process termed radiolysis. Since water comprises 55–60% of the human body, the probability of radiolysis is quite high under the presence of ionizing radiation. In the process, water loses an electron and becomes highly reactive. Then through a three-step chain reaction, water is sequentially converted to hydroxyl radical (•OH), hydrogen peroxide (H2O2), superoxide radical (•O−
2), and ultimately oxygen (O2).
The hydroxyl radical is extremely reactive and immediately removes electrons from any molecule in its path, turning that molecule into a free radical and thus propagating a chain reaction. However, hydrogen peroxide is actually more damaging to DNA than the hydroxyl radical, since the lower reactivity of hydrogen peroxide provides enough time for the molecule to travel into the nucleus of the cell, subsequently reacting with macromolecules such as DNA.
In plants, the production of ROS occurs during events of abiotic stress that lead to a reduction or interruption of metabolic activity. For example, the increase in temperature, drought are factors that limit the availability of CO2 due to stomatal closure, increasing the production of ROS, such as O2·- and 1O2 in chloroplasts.[25][26] The production of 1O2 in chloroplasts can cause reprogramming of the expression of nucleus genes leading to chlorosis and programmed cell death.[26] In cases of biotic stress, the generation of ROS occurs quickly and weakly initially and then becomes more solid and lasting.[27] The first phase of ROS accumulation is associated with plant infection and is probably independent of the synthesis of new ROS-generating enzymes. However, the second phase of ROS accumulation is associated only with infection by non-virulent pathogens and is an induced response dependent on increased mRNA transcription encoding enzymes.
Antioxidant enzymes
Superoxide dismutase
Superoxide dismutases (SOD) are a class of enzymes that catalyzes the dismutation of superoxide into oxygen and hydrogen peroxide. As such, they are an important antioxidant defense in nearly all cells exposed to oxygen. In mammals and most chordates, three forms of superoxide dismutase are present. SOD1 is located primarily in the cytoplasm, SOD2 in the mitochondria and SOD3 is extracellular. The first is a dimer (consists of two units), while the others are tetramers (four subunits). SOD1 and SOD3 contain copper and zinc ions, while SOD2 has a manganese ion in its reactive centre. The genes are located on chromosomes 21, 6, and 4, respectively (21q22.1, 6q25.3 and 4p15.3-p15.1).
The SOD-catalysed dismutation of superoxide may be written with the following half-reactions:
- M(n+1)+ − SOD + O−
2 → Mn+ − SOD + O2 - Mn+ − SOD + O−
2 + 2H+ → M(n+1)+ − SOD + H2O2.
where M = Cu (n = 1); Mn (n = 2); Fe (n = 2); Ni (n = 2). In this reaction the oxidation state of the metal cation oscillates between n and n + 1.
Catalase, which is concentrated in peroxisomes located next to mitochondria, reacts with the hydrogen peroxide to catalyze the formation of water and oxygen. Glutathione peroxidase reduces hydrogen peroxide by transferring the energy of the reactive peroxides to a sulfur-containing dipeptide called glutathione. The sulfur contained in these enzymes acts as the reactive center, carrying reactive electrons from the peroxide to the glutathione. Peroxiredoxins also degrade H2O2, within the mitochondria, cytosol, and nucleus.
- 2 H2O2 → 2 H2O + O2 (catalase)
- 2GSH + H2O2 → GS–SG + 2H2O (glutathione peroxidase)
Singlet oxygen
Another type of reactive oxygen species is singlet oxygen (1O2) which is produced for example as byproduct of photosynthesis in plants. In the presence of light and oxygen, photosensitizers such as chlorophyll may convert triplet (3O2) to singlet oxygen:[28]
Singlet oxygen is highly reactive, especially with organic compounds that contain double bonds. The resulting damage caused by singlet oxygen reduces the photosynthetic efficiency of chloroplasts. In plants exposed to excess light, the increased production of singlet oxygen can result in cell death.[28] Various substances such as carotenoids, tocopherols and plastoquinones contained in chloroplasts quench singlet oxygen and protect against its toxic effects. In addition to direct toxicity, singlet oxygen acts a signaling molecule.[28] Oxidized products of β-carotene arising from the presence of singlet oxygen act as second messengers that can either protect against singlet oxygen induced toxicity or initiate programmed cell death. Levels of jasmonate play a key role in the decision between cell acclimation or cell death in response to elevated levels of this reactive oxygen species.[28]
Damaging effects
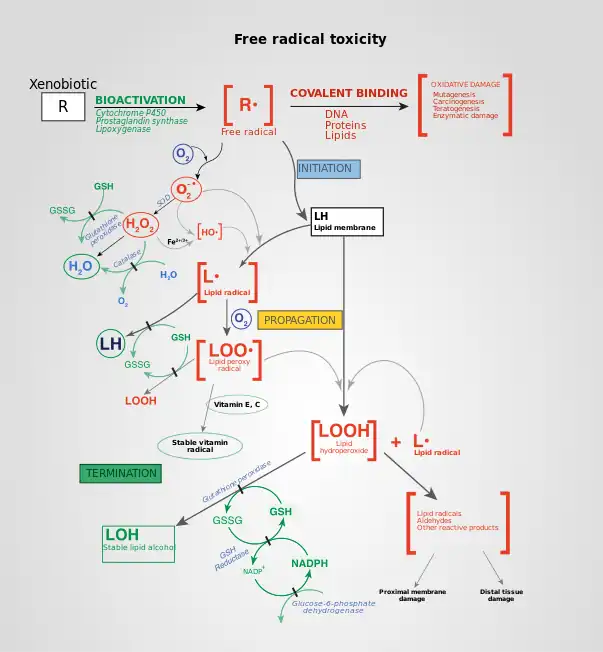
Effects of ROS on cell metabolism are well documented in a variety of species.[11] These include not only roles in apoptosis (programmed cell death) but also positive effects such as the induction of host defence[29][30]genes and mobilization of ion transporters. This implicates them in control of cellular function. In particular, platelets involved in wound repair and blood homeostasis release ROS to recruit additional platelets to sites of injury. These also provide a link to the adaptive immune system via the recruitment of leukocytes.
Reactive oxygen species are implicated in cellular activity to a variety of inflammatory responses including cardiovascular disease. They may also be involved in hearing impairment via cochlear damage induced by elevated sound levels, in ototoxicity of drugs such as cisplatin, and in congenital deafness in both animals and humans. ROS are also implicated in mediation of apoptosis or programmed cell death and ischaemic injury. Specific examples include stroke and heart attack.
In general, harmful effects of reactive oxygen species on the cell are most often:[31]
- damage of DNA or RNA
- oxidations of polyunsaturated fatty acids in lipids (lipid peroxidation)
- oxidations of amino acids in proteins
- oxidative deactivation of specific enzymes by oxidation of co-factors
Pathogen response
When a plant recognizes an attacking pathogen, one of the first induced reactions is to rapidly produce superoxide (O−
2) or hydrogen peroxide (H
2O
2) to strengthen the cell wall. This prevents the spread of the pathogen to other parts of the plant, essentially forming a net around the pathogen to restrict movement and reproduction.
In the mammalian host, ROS is induced as an antimicrobial defense.[20] To highlight the importance of this defense, individuals with chronic granulomatous disease who have deficiencies in generating ROS, are highly susceptible to infection by a broad range of microbes including Salmonella enterica, Staphylococcus aureus, Serratia marcescens, and Aspergillus spp.
Studies on the homeostasis of the Drosophila melanogaster’s intestines have shown the production of ROS as a key component of the immune response in the gut of the fly. ROS acts both as a bactericide, damaging the bacterial DNA, RNA and proteins, as well as a signalling molecule that induces repair mechanisms of the epithelium.[32] The uracil released by microorganism triggers the production and activity of Duox, the ROS-producing enzyme in the intestine. Duox activity is induced according to the level of uracil in the gut; under basal conditions, it is down-regulated by the protein kinase MkP3. The tight regulation of Duox avoids excessive production of ROS and facilitates differentiation between benign and damage-inducing microorganisms in the gut.[33]
The manner in which ROS defends the host from invading microbe is not fully understood. One of the more likely modes of defense is damage to microbial DNA. Studies using Salmonella demonstrated that DNA repair mechanisms were required to resist killing by ROS. A role for ROS in antiviral defense mechanisms has been demonstrated via Rig-like helicase-1 and mitochondrial antiviral signaling protein. Increased levels of ROS potentiate signaling through this mitochondria-associated antiviral receptor to activate interferon regulatory factor (IRF)-3, IRF-7, and nuclear factor kappa B (NF-κB), resulting in an antiviral state.[34] Respiratory epithelial cells induce mitochondrial ROS in response to influenza infection. This induction of ROS led to the induction of type III interferon and the induction of an antiviral state, limiting viral replication.[35] In host defense against mycobacteria, ROS play a role, although direct killing is likely not the key mechanism; rather, ROS likely affect ROS-dependent signalling controls, such as cytokine production, autophagy, and granuloma formation.[36][37]
Reactive oxygen species are also implicated in activation, anergy and apoptosis of T cells.[38]
Oxidative damage
In aerobic organisms the energy needed to fuel biological functions is produced in the mitochondria via the electron transport chain. Reactive oxygen species (ROS) with the potential to cause cellular damage are produced along with the release of energy. ROS can damage lipids, DNA, RNA, and proteins, which, in theory, contributes to the physiology of aging.
ROS are produced as a normal product of cellular metabolism. In particular, one major contributor to oxidative damage is hydrogen peroxide (H2O2), which is converted from superoxide that leaks from the mitochondria. Catalase and superoxide dismutase ameliorate the damaging effects of hydrogen peroxide and superoxide, respectively, by converting these compounds into oxygen and hydrogen peroxide (which is later converted to water), resulting in the production of benign molecules. However, this conversion is not 100% efficient, and residual peroxides persist in the cell. While ROS are produced as a product of normal cellular functioning, excessive amounts can cause deleterious effects.[39]
Impairment of cognitive function
Memory capabilities decline with age, evident in human degenerative diseases such as Alzheimer's disease, which is accompanied by an accumulation of oxidative damage. Current studies demonstrate that the accumulation of ROS can decrease an organism's fitness because oxidative damage is a contributor to senescence. In particular, the accumulation of oxidative damage may lead to cognitive dysfunction, as demonstrated in a study in which old rats were given mitochondrial metabolites and then given cognitive tests. Results showed that the rats performed better after receiving the metabolites, suggesting that the metabolites reduced oxidative damage and improved mitochondrial function.[40] Accumulating oxidative damage can then affect the efficiency of mitochondria and further increase the rate of ROS production.[41] The accumulation of oxidative damage and its implications for aging depends on the particular tissue type where the damage is occurring. Additional experimental results suggest that oxidative damage is responsible for age-related decline in brain functioning. Older gerbils were found to have higher levels of oxidized protein in comparison to younger gerbils. Treatment of old and young mice with a spin trapping compound caused a decrease in the level of oxidized proteins in older gerbils but did not have an effect on younger gerbils. In addition, older gerbils performed cognitive tasks better during treatment but ceased functional capacity when treatment was discontinued, causing oxidized protein levels to increase. This led researchers to conclude that oxidation of cellular proteins is potentially important for brain function.[42]
Cause of aging
According to the free radical theory of aging, oxidative damage initiated by reactive oxygen species is a major contributor to the functional decline that is characteristic of aging. While studies in invertebrate models indicate that animals genetically engineered to lack specific antioxidant enzymes (such as SOD), in general, show a shortened lifespan (as one would expect from the theory), the converse manipulation, increasing the levels of antioxidant enzymes, has yielded inconsistent effects on lifespan (though some studies in Drosophila do show that lifespan can be increased by the overexpression of MnSOD or glutathione biosynthesizing enzymes). Also contrary to this theory, deletion of mitochondrial SOD2 can extend lifespan in Caenorhabditis elegans.[43]
In mice, the story is somewhat similar. Deleting antioxidant enzymes, in general, yields shorter lifespan, although overexpression studies have not (with some exceptions) consistently extended lifespan.[44] Study of a rat model of premature aging found increased oxidative stress, reduced antioxidant enzyme activity and substantially greater DNA damage in the brain neocortex and hippocampus of the prematurely aged rats than in normally aging control rats.[45] The DNA damage 8-OHdG is a product of ROS interaction with DNA. Numerous studies have shown that 8-OHdG increases with age[46] (see DNA damage theory of aging).
Male infertility
Exposure of spermatozoa to oxidative stress is a major causative agent of male infertility.[47] Sperm DNA fragmentation, caused by oxidative stress, appears to be an important factor in the etiology of male infertility.[48] A high level of the oxidative DNA damage 8-OHdG is associated with abnormal spermatozoa and male infertility.[49]
Cancer
ROS are constantly generated and eliminated in the biological system and are required to drive regulatory pathways.[50] Under normal physiological conditions, cells control ROS levels by balancing the generation of ROS with their elimination by scavenging systems. But under oxidative stress conditions, excessive ROS can damage cellular proteins, lipids and DNA, leading to fatal lesions in the cell that contribute to carcinogenesis.
Cancer cells exhibit greater ROS stress than normal cells do, partly due to oncogenic stimulation, increased metabolic activity and mitochondrial malfunction. ROS is a double-edged sword. On one hand, at low levels, ROS facilitates cancer cell survival since cell-cycle progression driven by growth factors and receptor tyrosine kinases (RTK) require ROS for activation[51] and chronic inflammation, a major mediator of cancer, is regulated by ROS. On the other hand, a high level of ROS can suppress tumor growth through the sustained activation of cell-cycle inhibitor[52][53] and induction of cell death as well as senescence by damaging macromolecules. In fact, most of the chemotherapeutic and radiotherapeutic agents kill cancer cells by augmenting ROS stress.[54][55] The ability of cancer cells to distinguish between ROS as a survival or apoptotic signal is controlled by the dosage, duration, type, and site of ROS production. Modest levels of ROS are required for cancer cells to survive, whereas excessive levels kill them.
Metabolic adaptation in tumours balances the cells' need for energy with equally important need for macromolecular building blocks and tighter control of redox balance. As a result, production of NADPH is greatly enhanced, which functions as a cofactor to provide reducing power in many enzymatic reactions for macromolecular biosynthesis and at the same time rescuing the cells from excessive ROS produced during rapid proliferation. Cells counterbalance the detrimental effects of ROS by producing antioxidant molecules, such as reduced glutathione (GSH) and thioredoxin (TRX), which rely on the reducing power of NADPH to maintain their activities.[56]
Most risk factors associated with cancer interact with cells through the generation of ROS. ROS then activate various transcription factors such as nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), activator protein-1 (AP-1), hypoxia-inducible factor-1α and signal transducer and activator of transcription 3 (STAT3), leading to expression of proteins that control inflammation; cellular transformation; tumor cell survival; tumor cell proliferation; and invasion, agiogenesis as well as metastasis. And ROS also control the expression of various tumor suppressor genes such as p53, retinoblastoma gene (Rb), and phosphatase and tensin homolog (PTEN).[57]
Carcinogenesis
ROS-related oxidation of DNA is one of the main causes of mutations, which can produce several types of DNA damage, including non-bulky (8-oxoguanine and formamidopyrimidine) and bulky (cyclopurine and etheno adducts) base modifications, abasic sites, non-conventional single-strand breaks, protein-DNA adducts, and intra/interstrand DNA crosslinks.[58] It has been estimated that endogenous ROS produced via normal cell metabolism modify approximately 20,000 bases of DNA per day in a single cell. 8-oxoguanine is the most abundant among various oxidized nitrogeneous bases observed. During DNA replication, DNA polymerase mispairs 8-oxoguanine with adenine, leading to a G→T transversion mutation. The resulting genomic instability directly contributes to carcinogenesis. Cellular transformation leads to cancer and interaction of atypical PKC-ζ isoform with p47phox controls ROS production and transformation from apoptotic cancer stem cells through blebbishield emergency program.[59][60]
Cell proliferation
Uncontrolled proliferation is a hallmark of cancer cells. Both exogenous and endogenous ROS have been shown to enhance proliferation of cancer cells. The role of ROS in promoting tumor proliferation is further supported by the observation that agents with potential to inhibit ROS generation can also inhibit cancer cell proliferation.[57] Although ROS can promote tumor cell proliferation, a great increase in ROS has been associated with reduced cancer cell proliferation by induction of G2/M cell cycle arrest; increased phosphorylation of ataxia telangiectasia mutated (ATM), checkpoint kinase 1 (Chk 1), Chk 2; and reduced cell division cycle 25 homolog c (CDC25).[61]
Cell death
A cancer cell can die in three ways: apoptosis, necrosis, and autophagy. Excessive ROS can induce apoptosis through both the extrinsic and intrinsic pathways.[62] In the extrinsic pathway of apoptosis, ROS are generated by Fas ligand as an upstream event for Fas activation via phosphorylation, which is necessary for subsequent recruitment of Fas-associated protein with death domain and caspase 8 as well as apoptosis induction.[57] In the intrinsic pathway, ROS function to facilitate cytochrome c release by activating pore-stabilizing proteins (Bcl-2 and Bcl-xL) as well as inhibiting pore-destabilizing proteins (Bcl-2-associated X protein, Bcl-2 homologous antagonist/killer).[63] The intrinsic pathway is also known as the caspase cascade and is induced through mitochondrial damage which triggers the release of cytochrome c. DNA damage, oxidative stress, and loss of mitochondrial membrane potential lead to the release of the pro-apoptotic proteins mentioned above stimulating apoptosis.[64] Mitochondrial damage is closely linked to apoptosis and since mitochondria are easily targeted there is potential for cancer therapy.[65]
The cytotoxic nature of ROS is a driving force behind apoptosis, but in even higher amounts, ROS can result in both apoptosis and necrosis, a form of uncontrolled cell death, in cancer cells.[66]
Numerous studies have shown the pathways and associations between ROS levels and apoptosis, but a newer line of study has connected ROS levels and autophagy.[67] ROS can also induce cell death through autophagy, which is a self-catabolic process involving sequestration of cytoplasmic contents (exhausted or damaged organelles and protein aggregates) for degradation in lysosomes.[68] Therefore, autophagy can also regulate the cell's health in times of oxidative stress. Autophagy can be induced by ROS levels through many pathways in the cell in an attempt to dispose of harmful organelles and prevent damage, such as carcinogens, without inducing apoptosis.[69] Autophagic cell death can be prompted by the over expression of autophagy where the cell digests too much of itself in an attempt to minimize the damage and can no longer survive. When this type of cell death occurs, an increase or loss of control of autophagy regulating genes is commonly co-observed.[70] Thus, once a more in-depth understanding of autophagic cell death is attained and its relation to ROS, this form of programmed cell death may serve as a future cancer therapy. Autophagy and apoptosis are distinct mechanisms for cell death brought on by high levels of ROS. Aautophagy and apoptosis, however, rarely act through strictly independent pathways. There is a clear connection between ROS and autophagy and a correlation seen between excessive amounts of ROS leading to apoptosis.[69] The depolarization of the mitochondrial membrane is also characteristic of the initiation of autophagy. When mitochondria are damaged and begin to release ROS, autophagy is initiated to dispose of the damaging organelle. If a drug targets mitochondria and creates ROS, autophagy may dispose of so many mitochondria and other damaged organelles that the cell is no longer viable. The extensive amount of ROS and mitochondrial damage may also signal for apoptosis. The balance of autophagy within the cell and the crosstalk between autophagy and apoptosis mediated by ROS is crucial for a cell's survival. This crosstalk and connection between autophagy and apoptosis could be a mechanism targeted by cancer therapies or used in combination therapies for highly resistant cancers.
Tumor cell invasion, angiogenesis and metastasis
After growth factor stimulation of RTKs, ROS can trigger activation of signaling pathways involved in cell migration and invasion such as members of the mitogen activated protein kinase (MAPK) family – extracellular regulated kinase (ERK), c-jun NH-2 terminal kinase (JNK) and p38 MAPK. ROS can also promote migration by augmenting phosphorylation of the focal adhesion kinase (FAK) p130Cas and paxilin.[71]
Both in vitro and in vivo, ROS have been shown to induce transcription factors and modulate signaling molecules involved in angiogenesis (MMP, VEGF) and metastasis (upregulation of AP-1, CXCR4, AKT and downregulation of PTEN).[57]
Chronic inflammation and cancer
Experimental and epidemiologic research over the past several years has indicated close associations among ROS, chronic inflammation, and cancer.[57] ROS induces chronic inflammation by the induction of COX-2, inflammatory cytokines (TNFα, interleukin 1 (IL-1), IL-6), chemokines (IL-8, CXCR4) and pro-inflammatory transcription factors (NF-κB).[57] These chemokines and chemokine receptors, in turn, promote invasion and metastasis of various tumor types.
Cancer therapy
Both ROS-elevating and ROS-eliminating strategies have been developed with the former being predominantly used. Cancer cells with elevated ROS levels depend heavily on the antioxidant defense system. ROS-elevating drugs further increase cellular ROS stress level, either by direct ROS-generation (e.g. motexafin gadolinium, elesclomol) or by agents that abrogate the inherent antioxidant system such as SOD inhibitor (e.g. ATN-224, 2-methoxyestradiol) and GSH inhibitor (e.g. PEITC, buthionine sulfoximine (BSO)). The result is an overall increase in endogenous ROS, which when above a cellular tolerability threshold, may induce cell death.[72] On the other hand, normal cells appear to have, under lower basal stress and reserve, a higher capacity to cope with additional ROS-generating insults than cancer cells do.[73] Therefore, the elevation of ROS in all cells can be used to achieve the selective killing of cancer cells.
Radiotherapy also relies on ROS toxicity to eradicate tumor cells. Radiotherapy uses X-rays, γ-rays as well as heavy particle radiation such as protons and neutrons to induce ROS-mediated cell death and mitotic failure.[57]
Due to the dual role of ROS, both prooxidant and antioxidant-based anticancer agents have been developed. However, modulation of ROS signaling alone seems not to be an ideal approach due to adaptation of cancer cells to ROS stress, redundant pathways for supporting cancer growth and toxicity from ROS-generating anticancer drugs. Combinations of ROS-generating drugs with pharmaceuticals that can break the redox adaptation could be a better strategy for enhancing cancer cell cytotoxicity.[57]
James Watson[74] and others[75] have proposed that lack of intracellular ROS due to a lack of physical exercise may contribute to the malignant progression of cancer, because spikes of ROS are needed to correctly fold proteins in the endoplasmatic reticulum and low ROS levels may thus aspecifically hamper the formation of tumor suppressor proteins.[75] Since physical exercise induces temporary spikes of ROS, this may explain why physical exercise is beneficial for cancer patient prognosis.[76] Moreover, high inducers of ROS such as 2-deoxy-D-glucose and carbohydrate-based inducers of cellular stress induce cancer cell death more potently because they exploit the cancer cell's high avidity for sugars.[77]
Positive role of ROS in memory
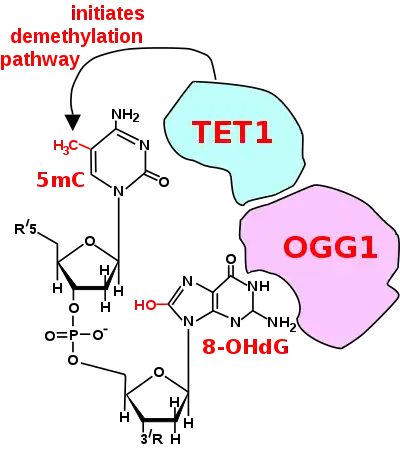
Two reviews[80][81] summarize the large body of evidence, reported largely between 1996 and 2011, for the critical and essential role of ROS in memory formation. An additional body of evidence indicates that both the formation and storage of memory depend on epigenetic modifications in neurons, including alterations in neuronal DNA methylation.[82][83] The two bodies of information on memory formation appear to be connected in 2016 by the work of Zhou et al.,[78] who showed that ROS have a central role in epigenetic DNA demethylation.
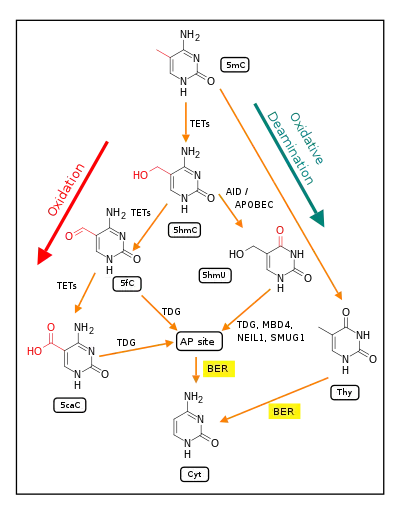
In mammalian nuclear DNA, a methyl group can be added, by a DNA methyltransferase, to the 5th carbon of cytosine to form 5mC (see red methyl group added to form 5mC near the top of the first figure). The DNA methyltransferases most often form 5mC within the dinucleotide sequence "cytosine-phosphate-guanine" to form 5mCpG. This addition is a major type of epigenetic alteration and it can silence gene expression. Methylated cytosine can also be demethylated, an epigenetic alteration that can increase the expression of a gene. A major enzyme involved in demethylating 5mCpG is TET1. However, TET1 is only able to act on 5mCpG if an ROS has first acted on the guanine to form 8-hydroxy-2'-deoxyguanosine (8-OHdG), resulting in a 5mCp-8-OHdG dinucleotide (see first figure).[78] However, TET1 is only able to act on the 5mC part of the dinucleotide when the base excision repair enzyme OGG1 binds to the 8-OHdG lesion without immediate excision. Adherence of OGG1 to the 5mCp-8-OHdG site recruits TET1 and TET1 then oxidizes the 5mC adjacent to 8-OHdG, as shown in the first figure, initiating a demethylation pathway shown in the second figure.
In 2016 Halder et al.[84] using mice, and in 2017 Duke et al.[85] using rats, subjected the rodents to contextual fear conditioning, causing an especially strong long-term memory to form. At 24 hours after the conditioning, in the hippocampus of rats, the expression of 1,048 genes was down-regulated (usually associated with hypermethylated gene promoters) and the expression of 564 genes was up-regulated (often associated with hypomethylated gene promoters). At 24 hours after training, 9.2% of the genes in the rat genome of hippocampus neurons were differentially methylated. However while the hippocampus is essential for learning new information it does not store information itself. In the mouse experiments of Halder, 1,206 differentially methylated genes were seen in the hippocampus one hour after contextual fear conditioning but these were reversed and not seen after four weeks. In contrast with the absence of long-term methylation changes in the hippocampus, substantial differential methylation could be detected in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex of mice four weeks after contextual fear conditioning.
The thousands of CpG sites being demethylated during memory formation depend on ROS in an initial step. The altered protein expression in neurons, controlled in part by ROS-dependent demethylation of CpG sites in gene promoters within neuron DNA, are central to memory formation.[86]
See also
- Antioxidant effect of polyphenols and natural phenols
- Iodide
- Melanin
- Mitohormesis
- Oxidative stress
- Oxygen toxicity
- Pro-oxidant
- Reactive nitrogen species
- Reactive sulfur species
- Reactive carbonyl species
- Reactive oxygen species production in marine microalgae
References
- ↑ Hayyan M, Hashim MA, AlNashef IM (March 2016). "Superoxide Ion: Generation and Chemical Implications". Chemical Reviews. 116 (5): 3029–85. doi:10.1021/acs.chemrev.5b00407. PMID 26875845.
- 1 2 3 Turrens JF (October 2003). "Mitochondrial formation of reactive oxygen species". The Journal of Physiology. 552 (Pt 2): 335–44. doi:10.1113/jphysiol.2003.049478. PMC 2343396. PMID 14561818.
- 1 2 Devasagayam TP, Tilak JC, Boloor KK, Sane KS, Ghaskadbi SS, Lele RD (October 2004). "Free radicals and antioxidants in human health: current status and future prospects". The Journal of the Association of Physicians of India. 52: 794–804. PMID 15909857.
- ↑ Edreva, Aglika (2 April 2005). "Generation and scavenging of reactive oxygen species in chloroplasts: a submolecular approach". Agriculture, Ecosystems & Environment. 106 (2): 119–133. doi:10.1016/j.agee.2004.10.022. ISSN 0167-8809. Retrieved 3 November 2020.
- ↑ Herb M, Gluschko A, Schramm M (September 2021). "Reactive Oxygen Species: Not Omnipresent but Important in Many Locations". Frontiers in Cell and Developmental Biology. 9 (716406): 716406. doi:10.3389/fcell.2021.716406. PMC 8452931. PMID 34557488.
- ↑ Grant JJ, Loake GJ (September 2000). "Role of reactive oxygen intermediates and cognate redox signaling in disease resistance". Plant Physiology. 124 (1): 21–9. doi:10.1104/pp.124.1.21. PMC 1539275. PMID 10982418.
- 1 2 3 Edreva, Aglika (2 April 2005). "Generation and scavenging of reactive oxygen species in chloroplasts: a submolecular approach". Agriculture, Ecosystems & Environment. 106 (2): 119–133. doi:10.1016/j.agee.2004.10.022. ISSN 0167-8809.
- ↑ Sosa Torres ME, Saucedo-Vázquez JP, Kroneck PM (2015). "Chapter 1, Section 3 The dark side of dioxygen". In Kroneck PM, Torres ME (eds.). Sustaining Life on Planet Earth: Metalloenzymes Mastering Dioxygen and Other Chewy Gases. Metal Ions in Life Sciences. Vol. 15. Springer. pp. 1–12. doi:10.1007/978-3-319-12415-5_1. PMID 25707464.
- ↑ Mittler R (January 2017). "ROS Are Good". Trends in Plant Science. 22 (1): 11–19. doi:10.1016/j.tplants.2016.08.002. PMID 27666517.
- ↑ Novo E, Parola M (October 2008). "Redox mechanisms in hepatic chronic wound healing and fibrogenesis". Fibrogenesis & Tissue Repair. 1 (1): 5. doi:10.1186/1755-1536-1-5. PMC 2584013. PMID 19014652.
- 1 2 3 Muthukumar K, Nachiappan V (December 2010). "Cadmium-induced oxidative stress in Saccharomyces cerevisiae". Indian Journal of Biochemistry & Biophysics. 47 (6): 383–7. PMID 21355423.
- ↑ Dietz KJ (January 2016). "Thiol-Based Peroxidases and Ascorbate Peroxidases: Why Plants Rely on Multiple Peroxidase Systems in the Photosynthesizing Chloroplast?". Molecules and Cells. 39 (1): 20–5. doi:10.14348/molcells.2016.2324. PMC 4749869. PMID 26810073.
- ↑ Muller F (October 2000). "The nature and mechanism of superoxide production by the electron transport chain: Its relevance to aging". Journal of the American Aging Association. 23 (4): 227–53. doi:10.1007/s11357-000-0022-9. PMC 3455268. PMID 23604868.
- ↑ Han D, Williams E, Cadenas E (January 2001). "Mitochondrial respiratory chain-dependent generation of superoxide anion and its release into the intermembrane space". The Biochemical Journal. 353 (Pt 2): 411–6. doi:10.1042/0264-6021:3530411. PMC 1221585. PMID 11139407.
- ↑ Li X, Fang P, Mai J, Choi ET, Wang H, Yang XF (February 2013). "Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers". Journal of Hematology & Oncology. 6 (19): 19. doi:10.1186/1756-8722-6-19. PMC 3599349. PMID 23442817.
- ↑ Hanukoglu I, Rapoport R, Weiner L, Sklan D (September 1993). "Electron leakage from the mitochondrial NADPH-adrenodoxin reductase-adrenodoxin-P450scc (cholesterol side chain cleavage) system". Archives of Biochemistry and Biophysics. 305 (2): 489–98. doi:10.1006/abbi.1993.1452. PMID 8396893.
- ↑ Hanukoglu I (2006). "Antioxidant protective mechanisms against reactive oxygen species (ROS) generated by mitochondrial P450 systems in steroidogenic cells". Drug Metabolism Reviews. 38 (1–2): 171–96. doi:10.1080/03602530600570040. PMID 16684656. S2CID 10766948.
- ↑ Curtin JF, Donovan M, Cotter TG (July 2002). "Regulation and measurement of oxidative stress in apoptosis". Journal of Immunological Methods. 265 (1–2): 49–72. doi:10.1016/s0022-1759(02)00070-4. PMID 12072178.
- ↑ Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Morgan, David; Raff, Martin; Roberts, Keith; Walter, Peter (2014). Molecular Biology of the Cell (6 ed.). New York: Garland Science. p. 1025. ISBN 978-0-8153-4432-2.
- 1 2 Herb M, Schramm M (February 2021). "Functions of ROS in Macrophages and Antimicrobial Immunity". Antioxidants. 10 (2): 313. doi:10.3390/antiox10020313. PMC 7923022. PMID 33669824.
- ↑ Chen, Xinfeng; Song, Mengjia; Zhang, Bin; Zhang, Yi (July 28, 2016). "Reactive Oxygen Species Regulate T Cell Immune Response in the Tumor Microenvironment". Oxidative Medicine and Cellular Longevity. 2016: 1–10. doi:10.1155/2016/1580967. PMC 4980531. PMID 27547291.
- ↑ Huang H, Ullah F, Zhou DX, Yi M, Zhao Y (2019). "Mechanisms of ROS Regulation of Plant Development and Stress Responses". Frontiers in Plant Science. 10: 800. doi:10.3389/fpls.2019.00800. PMC 6603150. PMID 31293607.
- 1 2 Zhang, Suping; Weng, Jun; Pan, Jingxi; Tu, Tiecheng; Yao, Side; Xu, Chunhe (1 January 2003). "Study on the photo-generation of superoxide radicals in Photosystem II with EPR spin trapping techniques". Photosynthesis Research. 75 (1): 41–48. doi:10.1023/A:1022439009587. ISSN 1573-5079. PMID 16245092. S2CID 11724647.
- ↑ Cleland, Robyn E.; Grace, Stephen C. (3 September 1999). "Voltammetric detection of superoxide production by photosystem II". FEBS Letters. 457 (3): 348–352. doi:10.1016/S0014-5793(99)01067-4. PMID 10471806. S2CID 1122939.
- ↑ Baniulis D, Hasan SS, Stofleth JT, Cramer WA (December 2013). "Mechanism of enhanced superoxide production in the cytochrome b(6)f complex of oxygenic photosynthesis". Biochemistry. 52 (50): 8975–83. doi:10.1021/bi4013534. PMC 4037229. PMID 24298890.
- 1 2 Kleine T, Leister D (August 2016). "Retrograde signaling: Organelles go networking". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1857 (8): 1313–1325. doi:10.1016/j.bbabio.2016.03.017. PMID 26997501.
- ↑ Grant JJ, Loake GJ (September 2000). "Role of reactive oxygen intermediates and cognate redox signaling in disease resistance". Plant Physiology. 124 (1): 21–9. doi:10.1104/pp.124.1.21. PMC 1539275. PMID 10982418.
- 1 2 3 4 Laloi C, Havaux M (2015). "Key players of singlet oxygen-induced cell death in plants". Frontiers in Plant Science. 6: 39. doi:10.3389/fpls.2015.00039. PMC 4316694. PMID 25699067.
- ↑ Rada B, Leto TL (2008). "Oxidative innate immune defenses by Nox/Duox family NADPH oxidases". In Egesten A, Schmidt A, Herwald H (eds.). Trends in Innate Immunity. Contributions to Microbiology. Vol. 15. Basel: Karger. pp. 164–87. doi:10.1159/000136357. ISBN 978-3-8055-8548-4. PMC 2776633. PMID 18511861. — Review
- ↑ Conner GE, Salathe M, Forteza R (December 2002). "Lactoperoxidase and hydrogen peroxide metabolism in the airway". American Journal of Respiratory and Critical Care Medicine. 166 (12 Pt 2): S57-61. doi:10.1164/rccm.2206018. PMID 12471090.
- ↑ Brooker RJ (2011). Genetics: analysis and principles (4th ed.). McGraw-Hill Science. ISBN 978-0-07-352528-0.
- ↑ Buchon N, Broderick NA, Lemaitre B (September 2013). "Gut homeostasis in a microbial world: insights from Drosophila melanogaster". Nature Reviews. Microbiology. 11 (9): 615–26. doi:10.1038/nrmicro3074. PMID 23893105. S2CID 8129204.
- ↑ Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, et al. (May 2013). "Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila". Cell. 153 (4): 797–811. doi:10.1016/j.cell.2013.04.009. PMID 23663779.
- ↑ West AP et al 2011 Nature Reviews Immunology 11, 389–402
- ↑ Kim HJ, Kim CH, Ryu JH, Kim MJ, Park CY, Lee JM, et al. (November 2013). "Reactive oxygen species induce antiviral innate immune response through IFN-λ regulation in human nasal epithelial cells". American Journal of Respiratory Cell and Molecular Biology. 49 (5): 855–65. doi:10.1165/rcmb.2013-0003OC. PMC 5455605. PMID 23786562.
- ↑ Herb M, Gluschko A, Wiegmann K, Farid A, Wolf A, Utermöhlen O, Krut O, Krönke M, Schramm M (February 2019). "Mitochondrial reactive oxygen species enable proinflammatory signaling through disulfide linkage of NEMO". Science Signaling. 12 (568): eaar5926. doi:10.1126/scisignal.aar5926. PMID 30755476.
- ↑ Deffert C, Cachat J, Krause KH (August 2014). "Phagocyte NADPH oxidase, chronic granulomatous disease and mycobacterial infections". Cellular Microbiology. 16 (8): 1168–78. doi:10.1111/cmi.12322. PMID 24916152. S2CID 3489742.
- ↑ Belikov AV, Schraven B, Simeoni L (October 2015). "T cells and reactive oxygen species". Journal of Biomedical Science. 22: 85. doi:10.1186/s12929-015-0194-3. PMC 4608155. PMID 26471060.
- ↑ Patel RP, T Cornwell T, Darley-Usmar VM (1999). "The biochemistry of nitric oxide and peroxynitrite: implications for mitochondrial function". In Packer L, Cadenas E (eds.). Understanding the process of aging: the roles of mitochondria, free radicals, and antioxidants. New York, NY: Marcel Dekker. pp. 39–56. ISBN 0-8247-1723-6.
- ↑ Liu J, Head E, Gharib AM, Yuan W, Ingersoll RT, Hagen TM, et al. (February 2002). "Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-L-carnitine and/or R-alpha -lipoic acid". Proceedings of the National Academy of Sciences of the United States of America. 99 (4): 2356–61. Bibcode:2002PNAS...99.2356L. doi:10.1073/pnas.261709299. PMC 122369. PMID 11854529.
- ↑ Stadtman ER (August 1992). "Protein oxidation and aging". Science. 257 (5074): 1220–4. Bibcode:1992Sci...257.1220S. doi:10.1126/science.1355616. PMID 1355616.
- ↑ Carney JM, Starke-Reed PE, Oliver CN, Landum RW, Cheng MS, Wu JF, Floyd RA (May 1991). "Reversal of age-related increase in brain protein oxidation, decrease in enzyme activity, and loss in temporal and spatial memory by chronic administration of the spin-trapping compound N-tert-butyl-alpha-phenylnitrone". Proceedings of the National Academy of Sciences of the United States of America. 88 (9): 3633–6. Bibcode:1991PNAS...88.3633C. doi:10.1073/pnas.88.9.3633. PMC 51506. PMID 1673789.
- ↑ Van Raamsdonk JM, Hekimi S (February 2009). "Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans". PLOS Genetics. 5 (2): e1000361. doi:10.1371/journal.pgen.1000361. PMC 2628729. PMID 19197346.
- ↑ Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H (August 2007). "Trends in oxidative aging theories". Free Radical Biology & Medicine. 43 (4): 477–503. doi:10.1016/j.freeradbiomed.2007.03.034. PMID 17640558.
- ↑ Sinha JK, Ghosh S, Swain U, Giridharan NV, Raghunath M (June 2014). "Increased macromolecular damage due to oxidative stress in the neocortex and hippocampus of WNIN/Ob, a novel rat model of premature aging". Neuroscience. 269: 256–64. doi:10.1016/j.neuroscience.2014.03.040. PMID 24709042. S2CID 9934178.
- ↑ Bernstein H, Payne CM, Bernstein C, Garewal H, Dvorak K (2008). "Chapter 1: Cancer and aging as consequences of un-repaired DNA damage.". In Kimura H, Suzuki A (eds.). New Research on DNA Damages. New York: Nova Science Publishers, Inc. pp. 1–47. ISBN 978-1-60456-581-2. Archived from the original on 2014-10-25. Retrieved 2018-03-15.
 , but read only.
, but read only. - ↑ Aitken RJ, De Iuliis GN, Gibb Z, Baker MA (August 2012). "The Simmet lecture: new horizons on an old landscape--oxidative stress, DNA damage and apoptosis in the male germ line". Reproduction in Domestic Animals = Zuchthygiene. 47 Suppl 4: 7–14. doi:10.1111/j.1439-0531.2012.02049.x. PMID 22827344.
- ↑ Wright C, Milne S, Leeson H (June 2014). "Sperm DNA damage caused by oxidative stress: modifiable clinical, lifestyle and nutritional factors in male infertility". Reproductive Biomedicine Online. 28 (6): 684–703. doi:10.1016/j.rbmo.2014.02.004. PMID 24745838.
- ↑ Guz J, Gackowski D, Foksinski M, Rozalski R, Zarakowska E, Siomek A, et al. (2013). "Comparison of oxidative stress/DNA damage in semen and blood of fertile and infertile men". PLOS ONE. 8 (7): e68490. Bibcode:2013PLoSO...868490G. doi:10.1371/journal.pone.0068490. PMC 3709910. PMID 23874641.
- ↑ Dickinson BC, Chang CJ (July 2011). "Chemistry and biology of reactive oxygen species in signaling or stress responses". Nature Chemical Biology. 7 (8): 504–11. doi:10.1038/nchembio.607. PMC 3390228. PMID 21769097.
- ↑ Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, et al. (March 1997). "Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts". Science. 275 (5306): 1649–52. doi:10.1126/science.275.5306.1649. PMID 9054359. S2CID 19733670.
- ↑ Ramsey MR, Sharpless NE (November 2006). "ROS as a tumour suppressor?". Nature Cell Biology. 8 (11): 1213–5. doi:10.1038/ncb1106-1213. PMID 17077852. S2CID 21104991.
- ↑ Takahashi A, Ohtani N, Yamakoshi K, Iida S, Tahara H, Nakayama K, et al. (November 2006). "Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence". Nature Cell Biology. 8 (11): 1291–7. doi:10.1038/ncb1491. PMID 17028578. S2CID 8686894.
- ↑ Renschler MF (September 2004). "The emerging role of reactive oxygen species in cancer therapy". European Journal of Cancer. 40 (13): 1934–40. doi:10.1016/j.ejca.2004.02.031. PMID 15315800.
- ↑ Toler SM, Noe D, Sharma A (December 2006). "Selective enhancement of cellular oxidative stress by chloroquine: implications for the treatment of glioblastoma multiforme". Neurosurgical Focus. 21 (6): E10. doi:10.3171/foc.2006.21.6.1. PMID 17341043.
- ↑ Cairns RA, Harris IS, Mak TW (February 2011). "Regulation of cancer cell metabolism". Nature Reviews. Cancer. 11 (2): 85–95. doi:10.1038/nrc2981. PMID 21258394. S2CID 8891526.
- 1 2 3 4 5 6 7 8 Gupta SC, Hevia D, Patchva S, Park B, Koh W, Aggarwal BB (June 2012). "Upsides and downsides of reactive oxygen species for cancer: the roles of reactive oxygen species in tumorigenesis, prevention, and therapy". Antioxidants & Redox Signaling. 16 (11): 1295–322. doi:10.1089/ars.2011.4414. PMC 3324815. PMID 22117137.
- ↑ Waris G, Ahsan H (May 2006). "Reactive oxygen species: role in the development of cancer and various chronic conditions". Journal of Carcinogenesis. 5: 14. doi:10.1186/1477-3163-5-14. PMC 1479806. PMID 16689993.
- ↑ Jinesh GG, Taoka R, Zhang Q, Gorantla S, Kamat AM (April 2016). "Novel PKC-ζ to p47 phox interaction is necessary for transformation from blebbishields". Scientific Reports. 6: 23965. Bibcode:2016NatSR...623965J. doi:10.1038/srep23965. PMC 4819220. PMID 27040869.
- ↑ Jinesh GG, Kamat AM. Blebbishield emergency program: an apoptotic route to cellular transformation. Cell Death Differ. 2016 In Press.
- ↑ Ames BN (September 1983). "Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases". Science. 221 (4617): 1256–64. Bibcode:1983Sci...221.1256A. doi:10.1126/science.6351251. PMID 6351251.
- ↑ Ozben T (September 2007). "Oxidative stress and apoptosis: impact on cancer therapy". Journal of Pharmaceutical Sciences. 96 (9): 2181–96. doi:10.1002/jps.20874. PMID 17593552.
- ↑ Martindale JL, Holbrook NJ (July 2002). "Cellular response to oxidative stress: signaling for suicide and survival". Journal of Cellular Physiology. 192 (1): 1–15. doi:10.1002/jcp.10119. PMID 12115731.
- ↑ Maiuri MC, Zalckvar E, Kimchi A, Kroemer G (September 2007). "Self-eating and self-killing: crosstalk between autophagy and apoptosis". Nature Reviews. Molecular Cell Biology. 8 (9): 741–52. doi:10.1038/nrm2239. PMID 17717517. S2CID 3912801.
- ↑ Fulda S, Galluzzi L, Kroemer G (June 2010). "Targeting mitochondria for cancer therapy". Nature Reviews. Drug Discovery. 9 (6): 447–64. doi:10.1038/nrd3137. PMID 20467424. S2CID 14643750.
- ↑ Hampton MB, Orrenius S (September 1997). "Dual regulation of caspase activity by hydrogen peroxide: implications for apoptosis". FEBS Letters. 414 (3): 552–6. doi:10.1016/s0014-5793(97)01068-5. PMID 9323034. S2CID 41952954.
- ↑ Gibson SB (October 2010). "A matter of balance between life and death: targeting reactive oxygen species (ROS)-induced autophagy for cancer therapy". Autophagy. 6 (7): 835–7. doi:10.4161/auto.6.7.13335. PMID 20818163.
- ↑ Shrivastava A, Kuzontkoski PM, Groopman JE, Prasad A (July 2011). "Cannabidiol induces programmed cell death in breast cancer cells by coordinating the cross-talk between apoptosis and autophagy". Molecular Cancer Therapeutics. 10 (7): 1161–72. doi:10.1158/1535-7163.MCT-10-1100. PMID 21566064.
- 1 2 Scherz-Shouval R, Elazar Z (September 2007). "ROS, mitochondria and the regulation of autophagy". Trends in Cell Biology. 17 (9): 422–7. doi:10.1016/j.tcb.2007.07.009. PMID 17804237.
- ↑ Xie Z, Klionsky DJ (October 2007). "Autophagosome formation: core machinery and adaptations". Nature Cell Biology. 9 (10): 1102–9. doi:10.1038/ncb1007-1102. PMID 17909521. S2CID 26402002.
- ↑ Tochhawng L, Deng S, Pervaiz S, Yap CT (May 2013). "Redox regulation of cancer cell migration and invasion". Mitochondrion. 13 (3): 246–53. doi:10.1016/j.mito.2012.08.002. PMID 22960576.
- ↑ Schumacker PT (September 2006). "Reactive oxygen species in cancer cells: live by the sword, die by the sword". Cancer Cell. 10 (3): 175–6. doi:10.1016/j.ccr.2006.08.015. PMID 16959608.
- ↑ Trachootham D, Alexandre J, Huang P (July 2009). "Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach?". Nature Reviews. Drug Discovery. 8 (7): 579–91. doi:10.1038/nrd2803. PMID 19478820. S2CID 20697221.
- ↑ Watson JD (March 2014). "Type 2 diabetes as a redox disease". Lancet. 383 (9919): 841–3. doi:10.1016/s0140-6736(13)62365-x. PMID 24581668. S2CID 1076963.
- 1 2 Molenaar RJ, van Noorden CJ (September 2014). "Type 2 diabetes and cancer as redox diseases?". Lancet. 384 (9946): 853. doi:10.1016/s0140-6736(14)61485-9. PMID 25209484. S2CID 28902284.
- ↑ Irwin ML, Smith AW, McTiernan A, Ballard-Barbash R, Cronin K, Gilliland FD, et al. (August 2008). "Influence of pre- and postdiagnosis physical activity on mortality in breast cancer survivors: the health, eating, activity, and lifestyle study". Journal of Clinical Oncology. 26 (24): 3958–64. doi:10.1200/jco.2007.15.9822. PMC 2654316. PMID 18711185.
- ↑ Ndombera FT, VanHecke GC, Nagi S, Ahn YH (March 2016). "Carbohydrate-based inducers of cellular stress for targeting cancer cells". Bioorganic & Medicinal Chemistry Letters. 26 (5): 1452–6. doi:10.1016/j.bmcl.2016.01.063. PMID 26832785.
- 1 2 3 Zhou X, Zhuang Z, Wang W, He L, Wu H, Cao Y, et al. (September 2016). "OGG1 is essential in oxidative stress induced DNA demethylation". Cellular Signalling. 28 (9): 1163–71. doi:10.1016/j.cellsig.2016.05.021. PMID 27251462.
- ↑ Bayraktar G, Kreutz MR (2018). "The Role of Activity-Dependent DNA Demethylation in the Adult Brain and in Neurological Disorders". Frontiers in Molecular Neuroscience. 11: 169. doi:10.3389/fnmol.2018.00169. PMC 5975432. PMID 29875631.
- ↑ Massaad CA, Klann E (May 2011). "Reactive oxygen species in the regulation of synaptic plasticity and memory". Antioxidants & Redox Signaling. 14 (10): 2013–54. doi:10.1089/ars.2010.3208. PMC 3078504. PMID 20649473.
- ↑ Beckhauser TF, Francis-Oliveira J, De Pasquale R (2016). "Reactive Oxygen Species: Physiological and Physiopathological Effects on Synaptic Plasticity". Journal of Experimental Neuroscience. 10 (Suppl 1): 23–48. doi:10.4137/JEN.S39887. PMC 5012454. PMID 27625575.
- ↑ Day JJ, Sweatt JD (January 2011). "Epigenetic modifications in neurons are essential for formation and storage of behavioral memory". Neuropsychopharmacology. 36 (1): 357–8. doi:10.1038/npp.2010.125. PMC 3055499. PMID 21116250.
- ↑ Sweatt JD (October 2016). "Neural plasticity and behavior - sixty years of conceptual advances". Journal of Neurochemistry. 139 Suppl 2: 179–199. doi:10.1111/jnc.13580. PMID 26875778.
- ↑ Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, et al. (January 2016). "DNA methylation changes in plasticity genes accompany the formation and maintenance of memory". Nature Neuroscience. 19 (1): 102–10. doi:10.1038/nn.4194. PMC 4700510. PMID 26656643.
- ↑ Duke CG, Kennedy AJ, Gavin CF, Day JJ, Sweatt JD (July 2017). "Experience-dependent epigenomic reorganization in the hippocampus". Learning & Memory. 24 (7): 278–288. doi:10.1101/lm.045112.117. PMC 5473107. PMID 28620075.
- ↑ Day JJ, Sweatt JD (November 2010). "DNA methylation and memory formation". Nature Neuroscience. 13 (11): 1319–23. doi:10.1038/nn.2666. PMC 3130618. PMID 20975755.
Further reading
- Sen CK (2003). "The general case for redox control of wound repair". Wound Repair and Regeneration. 11 (6): 431–8. doi:10.1046/j.1524-475X.2003.11607.x. PMID 14617282. S2CID 40770160.
- Krötz F, Sohn HY, Gloe T, Zahler S, Riexinger T, Schiele TM, et al. (August 2002). "NAD(P)H oxidase-dependent platelet superoxide anion release increases platelet recruitment". Blood. 100 (3): 917–24. doi:10.1182/blood.V100.3.917. PMID 12130503.
- Pignatelli P, Pulcinelli FM, Lenti L, Gazzaniga PP, Violi F (January 1998). "Hydrogen peroxide is involved in collagen-induced platelet activation". Blood. 91 (2): 484–90. doi:10.1182/blood.V91.2.484. PMID 9427701.
- Guzik TJ, Korbut R, Adamek-Guzik T (December 2003). "Nitric oxide and superoxide in inflammation and immune regulation". Journal of Physiology and Pharmacology. 54 (4): 469–87. PMID 14726604.
External links
- Nobel laureate James Watson's novel hypothesis
- Antioxidants don't work, but no one wants to hear it.
- Archived 2016-09-27 at the Wayback Machine