Reverse transcriptase
| Reverse transcriptase (RNA-dependent DNA polymerase) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | RVT_1 | ||||||||
| Pfam | PF00078 | ||||||||
| Pfam clan | CL0027 | ||||||||
| InterPro | IPR000477 | ||||||||
| PROSITE | PS50878 | ||||||||
| SCOP2 | 1hmv / SCOPe / SUPFAM | ||||||||
| CDD | cd00304 | ||||||||
| |||||||||
| RNA-directed DNA polymerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 2.7.7.49 | ||||||||
| CAS no. | 9068-38-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible.[2][3][4]
Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genome, from which new RNA copies can be made via host-cell transcription. The same sequence of reactions is widely used in the laboratory to convert RNA to DNA for use in molecular cloning, RNA sequencing, polymerase chain reaction (PCR), or genome analysis.
History
Reverse transcriptases were discovered by Howard Temin at the University of Wisconsin–Madison in Rous sarcoma virions[5] and independently isolated by David Baltimore in 1970 at MIT from two RNA tumour viruses: murine leukemia virus and again Rous sarcoma virus.[6] For their achievements, they shared the 1975 Nobel Prize in Physiology or Medicine (with Renato Dulbecco).
Well-studied reverse transcriptases include:
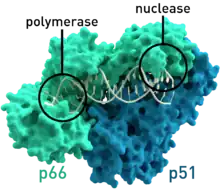
- HIV-1 reverse transcriptase from human immunodeficiency virus type 1 (PDB: 1HMV) has two subunits, which have respective molecular weights of 66 and 51 kDas.[7]
- M-MLV reverse transcriptase from the Moloney murine leukemia virus is a single 75 kDa monomer.[8]
- AMV reverse transcriptase from the avian myeloblastosis virus also has two subunits, a 63 kDa subunit and a 95 kDa subunit.[8]
- Telomerase reverse transcriptase that maintains the telomeres of eukaryotic chromosomes.[9]
Function in viruses
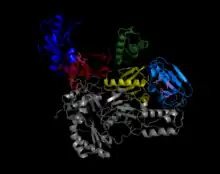
The enzymes are encoded and used by viruses that use reverse transcription as a step in the process of replication. Reverse-transcribing RNA viruses, such as retroviruses, use the enzyme to reverse-transcribe their RNA genomes into DNA, which is then integrated into the host genome and replicated along with it. Reverse-transcribing DNA viruses, such as the hepadnaviruses, can allow RNA to serve as a template in assembling and making DNA strands. HIV infects humans with the use of this enzyme. Without reverse transcriptase, the viral genome would not be able to incorporate into the host cell, resulting in failure to replicate.
Process of reverse transcription or retrotranscription
Reverse transcriptase creates double-stranded DNA from an RNA template.
In virus species with reverse transcriptase lacking DNA-dependent DNA polymerase activity, creation of double-stranded DNA can possibly be done by host-encoded DNA polymerase δ, mistaking the viral DNA-RNA for a primer and synthesizing a double-stranded DNA by a similar mechanism as in primer removal, where the newly synthesized DNA displaces the original RNA template.
The process of reverse transcription, also called retrotranscription or retrotras, is extremely error-prone, and it is during this step that mutations may occur. Such mutations may cause drug resistance.
Retroviral reverse transcription
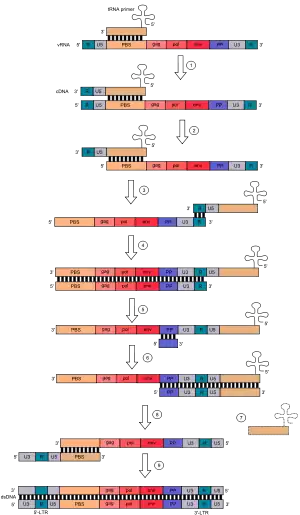
Retroviruses, also referred to as class VI ssRNA-RT viruses, are RNA reverse-transcribing viruses with a DNA intermediate. Their genomes consist of two molecules of positive-sense single-stranded RNA with a 5' cap and 3' polyadenylated tail. Examples of retroviruses include the human immunodeficiency virus (HIV) and the human T-lymphotropic virus (HTLV). Creation of double-stranded DNA occurs in the cytosol[10] as a series of these steps:
- Lysyl tRNA acts as a primer and hybridizes to a complementary part of the virus RNA genome called the primer binding site or PBS.
- Reverse transcriptase then adds DNA nucleotides onto the 3' end of the primer, synthesizing DNA complementary to the U5 (non-coding region) and R region (a direct repeat found at both ends of the RNA molecule) of the viral RNA.
- A domain on the reverse transcriptase enzyme called RNAse H degrades the U5 and R regions on the 5’ end of the RNA.
- The tRNA primer then "jumps" to the 3’ end of the viral genome, and the newly synthesised DNA strands hybridizes to the complementary R region on the RNA.
- The complementary DNA (cDNA) added in (2) is further extended.
- The majority of viral RNA is degraded by RNAse H, leaving only the PP sequence.
- Synthesis of the second DNA strand begins, using the remaining PP fragment of viral RNA as a primer.
- The tRNA primer leaves and a "jump" happens. The PBS from the second strand hybridizes with the complementary PBS on the first strand.
- Both strands are extended to form a complete double-stranded DNA copy of the original viral RNA genome, which can then be incorporated into the host's genome by the enzyme integrase.
Creation of double-stranded DNA also involves strand transfer, in which there is a translocation of short DNA product from initial RNA-dependent DNA synthesis to acceptor template regions at the other end of the genome, which are later reached and processed by the reverse transcriptase for its DNA-dependent DNA activity.[11]
Retroviral RNA is arranged in 5’ terminus to 3’ terminus. The site where the primer is annealed to viral RNA is called the primer-binding site (PBS). The RNA 5’end to the PBS site is called U5, and the RNA 3’ end to the PBS is called the leader. The tRNA primer is unwound between 14 and 22 nucleotides and forms a base-paired duplex with the viral RNA at PBS. The fact that the PBS is located near the 5’ terminus of viral RNA is unusual because reverse transcriptase synthesize DNA from 3’ end of the primer in the 5’ to 3’ direction (with respect to the newly synthesized DNA strand). Therefore, the primer and reverse transcriptase must be relocated to 3’ end of viral RNA. In order to accomplish this reposition, multiple steps and various enzymes including DNA polymerase, ribonuclease H(RNase H) and polynucleotide unwinding are needed.[12][13]
The HIV reverse transcriptase also has ribonuclease activity that degrades the viral RNA during the synthesis of cDNA, as well as DNA-dependent DNA polymerase activity that copies the sense cDNA strand into an antisense DNA to form a double-stranded viral DNA intermediate (vDNA).[14]
In cellular life
Self-replicating stretches of eukaryotic genomes known as retrotransposons utilize reverse transcriptase to move from one position in the genome to another via an RNA intermediate. They are found abundantly in the genomes of plants and animals. Telomerase is another reverse transcriptase found in many eukaryotes, including humans, which carries its own RNA template; this RNA is used as a template for DNA replication.[15]
Initial reports of reverse transcriptase in prokaryotes came as far back as 1971 in France (Beljanski et al., 1971a, 1972) and a few years later in the USSR (Romashchenko 1977[16]). These have since been broadly described as part of bacterial Retrons, distinct sequences that code for reverse transcriptase, and are used in the synthesis of msDNA. In order to initiate synthesis of DNA, a primer is needed. In bacteria, the primer is synthesized during replication.[17]
Valerian Dolja of Oregon State argues that viruses, due to their diversity, have played an evolutionary role in the development of cellular life, with reverse transcriptase playing a central role.[18]
Structure
The reverse transcriptase employs a "right hand" structure similar to that found in other viral nucleic acid polymerases.[19][20] In addition to the transcription function, retroviral reverse transcriptases have a domain belonging to the RNase H family, which is vital to their replication. By degrading the RNA template, it allows the other strand of DNA to be synthesized.[21] Some fragments from the digestion also serve as the primer for the DNA polymerase (either the same enzyme or a host protein), responsible for making the other (plus) strand.[19]
Replication fidelity
There are three different replication systems during the life cycle of a retrovirus. The first process is the reverse transcriptase synthesis of viral DNA from viral RNA, which then forms newly made complementary DNA strands. The second replication process occurs when host cellular DNA polymerase replicates the integrated viral DNA. Lastly, RNA polymerase II transcribes the proviral DNA into RNA, which will be packed into virions. Mutation can occur during one or all of these replication steps.[22]
Reverse transcriptase has a high error rate when transcribing RNA into DNA since, unlike most other DNA polymerases, it has no proofreading ability. This high error rate allows mutations to accumulate at an accelerated rate relative to proofread forms of replication. The commercially available reverse transcriptases produced by Promega are quoted by their manuals as having error rates in the range of 1 in 17,000 bases for AMV and 1 in 30,000 bases for M-MLV.[23]
Other than creating single-nucleotide polymorphisms, reverse transcriptases have also been shown to be involved in processes such as transcript fusions, exon shuffling and creating artificial antisense transcripts.[24][25] It has been speculated that this template switching activity of reverse transcriptase, which can be demonstrated completely in vivo, may have been one of the causes for finding several thousand unannotated transcripts in the genomes of model organisms.[26]
Template switching
Two RNA genomes are packaged into each retrovirus particle, but, after an infection, each virus generates only one provirus.[27] After infection, reverse transcription is accompanied by template switching between the two genome copies (copy choice recombination).[27] There are two models that suggest why RNA transcriptase switches templates. The first, the forced copy-choice model, proposes that reverse transcriptase changes the RNA template when it encounters a nick, implying that recombination is obligatory to maintaining virus genome integrity. The second, the dynamic choice model, suggests that reverse transcriptase changes templates when the RNAse function and the polymerase function are not in sync rate-wise, implying that recombination occurs at random and is not in response to genomic damage. A study by Rawson et al. supported both models of recombination.[27] From 5 to 14 recombination events per genome occur at each replication cycle.[28] Template switching (recombination) appears to be necessary for maintaining genome integrity and as a repair mechanism for salvaging damaged genomes.[29][27]
Applications
Antiviral drugs
As HIV uses reverse transcriptase to copy its genetic material and generate new viruses (part of a retrovirus proliferation circle), specific drugs have been designed to disrupt the process and thereby suppress its growth. Collectively, these drugs are known as reverse-transcriptase inhibitors and include the nucleoside and nucleotide analogues zidovudine (trade name Retrovir), lamivudine (Epivir) and tenofovir (Viread), as well as non-nucleoside inhibitors, such as nevirapine (Viramune).
Molecular biology
Reverse transcriptase is commonly used in research to apply the polymerase chain reaction technique to RNA in a technique called reverse transcription polymerase chain reaction (RT-PCR). The classical PCR technique can be applied only to DNA strands, but, with the help of reverse transcriptase, RNA can be transcribed into DNA, thus making PCR analysis of RNA molecules possible. Reverse transcriptase is used also to create cDNA libraries from mRNA. The commercial availability of reverse transcriptase greatly improved knowledge in the area of molecular biology, as, along with other enzymes, it allowed scientists to clone, sequence, and characterise RNA.
Reverse transcriptase has also been employed in insulin production. By inserting eukaryotic mRNA for insulin production along with reverse transcriptase into bacteria, the mRNA could be inserted into the prokaryote's genome. Large amounts of insulin can then be created, sidestepping the need to harvest pig pancreas and other such traditional sources. Directly inserting eukaryotic DNA into bacteria would not work because it carries introns, so would not translate successfully using the bacterial ribosomes. Processing in the eukaryotic cell during mRNA production removes these introns to provide a suitable template. Reverse transcriptase converts this edited RNA back into DNA so it could be incorporated in the genome.
See also
- cDNA library
- DNA polymerase
- msDNA
- Reverse transcribing virus
- RNA polymerase
- Telomerase
- Retrotransposon marker
References
- ↑ PDB: 3KLF; Tu X, Das K, Han Q, Bauman JD, Clark AD, Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, Hughes SH, Sarafianos SG, Arnold E (October 2010). "Structural basis of HIV-1 resistance to AZT by excision". Nature Structural & Molecular Biology. 17 (10): 1202–9. doi:10.1038/nsmb.1908. PMC 2987654. PMID 20852643.
- ↑ Crick F (August 1970). "Central dogma of molecular biology". Nature. 227 (5258): 561–3. Bibcode:1970Natur.227..561C. doi:10.1038/227561a0. PMID 4913914. S2CID 4164029.
- ↑ Sarkar S (1996). The Philosophy and History of Molecular Biology: New Perspectives. Dordrecht: Kluwer Academic Publishers. pp. 187–232.
- ↑ Danchin É, Pocheville A, Rey O, Pujol B, Blanchet S (2019). "Epigenetically facilitated mutational assimilation: epigenetics as a hub within the inclusive evolutionary synthesis". Biological Reviews. 94: 259–282. doi:10.1111/brv.12453. PMC 6378602. S2CID 67861162.
- ↑ Temin HM, Mizutani S (June 1970). "RNA-dependent DNA polymerase in virions of Rous sarcoma virus". Nature. 226 (5252): 1211–3. doi:10.1038/2261211a0. PMID 4316301. S2CID 4187764.
- ↑ Baltimore D (June 1970). "RNA-dependent DNA polymerase in virions of RNA tumour viruses". Nature. 226 (5252): 1209–11. doi:10.1038/2261209a0. PMID 4316300. S2CID 4222378.
- ↑ Ferris AL, Hizi A, Showalter SD, Pichuantes S, Babe L, Craik CS, Hughes SH (April 1990). "Immunologic and proteolytic analysis of HIV-1 reverse transcriptase structure" (PDF). Virology. 175 (2): 456–64. doi:10.1016/0042-6822(90)90430-y. PMID 1691562.
- 1 2 Konishi A, Yasukawa K, Inouye K (July 2012). "Improving the thermal stability of avian myeloblastosis virus reverse transcriptase α-subunit by site-directed mutagenesis" (PDF). Biotechnology Letters. 34 (7): 1209–15. doi:10.1007/s10529-012-0904-9. hdl:2433/157247. PMID 22426840. S2CID 207096569.
- ↑ Autexier C, Lue NF (June 2006). "The structure and function of telomerase reverse transcriptase". Annual Review of Biochemistry. 75 (1): 493–517. doi:10.1146/annurev.biochem.75.103004.142412. PMID 16756500.
- ↑ Bio-Medicine.org - Retrovirus Retrieved on 17 Feb, 2009
- ↑ Telesnitsky A, Goff SP (1993). "Strong-stop strand transfer during reverse transcription". In Skalka MA, Goff SP (eds.). Reverse transcriptase (1st ed.). New York: Cold Spring Harbor. p. 49. ISBN 978-0-87969-382-4.
- ↑ Bernstein A, Weiss R, Tooze J (1985). "RNA tumor viruses". Molecular Biology of Tumor Viruses (2nd ed.). Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory.
- ↑ Moelling K, Broecker F (April 2015). "The reverse transcriptase-RNase H: from viruses to antiviral defense". Annals of the New York Academy of Sciences. 1341 (1): 126–35. Bibcode:2015NYASA1341..126M. doi:10.1111/nyas.12668. PMID 25703292. S2CID 42378727.
- ↑ Kaiser GE (January 2008). "The Life Cycle of HIV". Doc Kaiser's Microbiology Home Page. Community College of Baltimore Count. Archived from the original on 2010-07-26.
- ↑ Krieger M, Scott MP, Matsudaira PT, Lodish HF, Darnell JE, Zipursky L, Kaiser C, Berk A (2004). Molecular cell biology. New York: W.H. Freeman and CO. ISBN 978-0-7167-4366-8.
- ↑ Romashchenko AG, et al. (1977). "Otdelenie ot preparatov DNK-polimeraz I RNK-zavisimoy DNK-polimeraz; oshistka i svoystva fermenta". Proceedings of the USSR Academy of Sciences. 233: 734–737.
- ↑ Hurwitz J, Leis JP (January 1972). "RNA-dependent DNA polymerase activity of RNA tumor viruses. I. Directing influence of DNA in the reaction". Journal of Virology. 9 (1): 116–29. doi:10.1128/JVI.9.1.116-129.1972. PMC 356270. PMID 4333538.
- ↑ Arnold C (17 July 2014). "Could Giant Viruses Be the Origin of Life on Earth?". National Geographic. Retrieved 29 May 2016.
- 1 2 Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E (January 2009). "Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition". Journal of Molecular Biology. 385 (3): 693–713. doi:10.1016/j.jmb.2008.10.071. PMC 2881421. PMID 19022262.
- ↑ Hansen JL, Long AM, Schultz SC (August 1997). "Structure of the RNA-dependent RNA polymerase of poliovirus". Structure. 5 (8): 1109–22. doi:10.1016/S0969-2126(97)00261-X. PMID 9309225.
- ↑ Schultz SJ, Champoux JJ (June 2008). "RNase H activity: structure, specificity, and function in reverse transcription". Virus Research. 134 (1–2): 86–103. doi:10.1016/j.virusres.2007.12.007. PMC 2464458. PMID 18261820.
- ↑ Bbenek K, Kunkel AT (1993). "The fidelity of retroviral reverse transcriptases". In Skalka MA, Goff PS (eds.). Reverse transcriptase. New York: Cold Spring Harbor Laboratory Press. p. 85. ISBN 978-0-87969-382-4.
- ↑ "Promega kit instruction manual" (PDF). 1999. Archived from the original (PDF) on 2006-11-21.
- ↑ Houseley J, Tollervey D (August 2010). "Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro". PLOS ONE. 5 (8): e12271. Bibcode:2010PLoSO...512271H. doi:10.1371/journal.pone.0012271. PMC 2923612. PMID 20805885.
- ↑ Zeng XC, Wang SX (June 2002). "Evidence that BmTXK beta-BmKCT cDNA from Chinese scorpion Buthus martensii Karsch is an artifact generated in the reverse transcription process". FEBS Letters. 520 (1–3): 183–4, author reply 185. doi:10.1016/S0014-5793(02)02812-0. PMID 12044895. S2CID 24619868.
- ↑ van Bakel H, Nislow C, Blencowe BJ, Hughes TR (2011). "Response to "The Reality of Pervasive Transcription"". PLOS Biology. 9 (7): e1001102. doi:10.1371/journal.pbio.1001102. PMC 3134445.
- 1 2 3 4 Rawson JM, Nikolaitchik OA, Keele BF, Pathak VK, Hu WS (November 2018). "Recombination is required for efficient HIV-1 replication and the maintenance of viral genome integrity". Nucleic Acids Research. 46 (20): 10535–10545. doi:10.1093/nar/gky910. PMC 6237782. PMID 30307534.
- ↑ Cromer D, Grimm AJ, Schlub TE, Mak J, Davenport MP (January 2016). "Estimating the in-vivo HIV template switching and recombination rate". AIDS (London, England). 30 (2): 185–92. doi:10.1097/QAD.0000000000000936. PMID 26691546. S2CID 20086739.
- ↑ Hu WS, Temin HM (November 1990). "Retroviral recombination and reverse transcription". Science. New York, N.Y. 250 (4985): 1227–33. Bibcode:1990Sci...250.1227H. doi:10.1126/science.1700865. PMID 1700865.
External links
- RNA+Transcriptase at the US National Library of Medicine Medical Subject Headings (MeSH)
- animation of reverse transcriptase action and three reverse transcriptase inhibitors
- Molecule of the month (September 2002) at the RCSB PDB
- HIV Replication 3D Medical Animation. (Nov 2008). Video by Boehringer Ingelheim.
- Goodsell DS. "Molecule of the Month: Reverse Transcriptase (Sep 2002)". Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). Retrieved 2013-01-13.
- Overview of all the structural information available in the PDB for UniProt: P03366 (Human immunodeficiency virus Reverse transcriptase) at the PDBe-KB.