Rickettsiaceae
| Rickettsiaceae | |
|---|---|
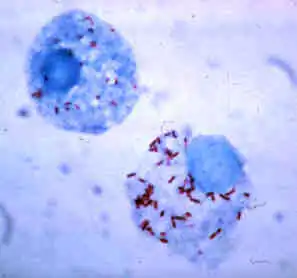 | |
| Rickettsia rickettsii (red dots) in the cell of a deer tick | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Proteobacteria |
| Class: | Alphaproteobacteria |
| Order: | Rickettsiales |
| Family: | Rickettsiaceae Pinkerton 1936 (Approved Lists 1980) |
| Tribes and genera | |
| |
The Rickettsiaceae are a family of bacteria. The genus Rickettsia is the most prominent genus within the family. From this family, the bacteria that eventually formed the mitochondrion (an organelle in eukaryotic cells) is believed to have originated. Most human pathogens in this family are in genus Rickettsia. They spend part of their lifecycle in the bodies of arthropods such as ticks or lice, and are then transmitted to humans or other mammals by the bite of the arthropod. It contains Gram-negative bacteria, very sensitive to environmental exposure, thus is adapted to obligate intracellular infection. Rickettsia rickettsii is considered the prototypical infectious organism in the group.
Genomics
Comparative genomic analysis has identified three proteins, RP030, RP187 and RP192, which are uniquely found in members of the family Rickettsiaceae and serve as molecular markers for this family.[2] In addition, conserved signature indels in a number of proteins including a four-amino-acid insert in transcription repair coupling factor Mfd, a 10-amino-acid insert in ribosomal protein L19, one-amino-acid inserts each in the FtsZ protein and the major sigma factor 70, and a one-amino-acid deletion in exonuclease VII protein that are specific for the Rickettsiaceae species have been identified.[3]
| Schematic ribosomal RNA phylogeny of Alphaproteobacteria | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| The cladogram of Rickettsidae has been inferred by Ferla et al. [4] from the comparison of 16S + 23S ribosomal RNA sequences. |
References
- 1 2 Vannini C, Boscaro V, Ferrantini F, Benken KA, Mironov TI, Schweikert M, Görtz H-D, Fokin SI, Sabaneyeva EV, Petroni G. (2014). "Flagellar Movement in Two Bacteria of the Family Rickettsiaceae: A Re-Evaluation of Motility in an Evolutionary Perspective". PLOS ONE. 9 (2): e87718. doi:10.1371/journal.pone.0087718. PMC 3914857. PMID 24505307.
{{cite journal}}: CS1 maint: uses authors parameter (link) - ↑ Gupta, R. S. and Mok, A. (2007). Phylogenomics and signature proteins for the alpha Proteobacteria and its main groups. BMC Microbiology. 7:106. doi:10.1186/1471-2180-7-106.
- ↑ Gupta, R. S. (2005). Protein signatures distinctive of alpha proteobacteria and its subgroups and a model for alpha proteobacterial evolution. Critical Reviews in Microbiology. 3:101-135. doi:10.1080/10408410590922393.
- ↑ Ferla MP, Thrash JC, Giovannoni SJ, Patrick WM (2013). "New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability". PLOS ONE. 8 (12): e83383. Bibcode:2013PLoSO...883383F. doi:10.1371/journal.pone.0083383. PMC 3859672. PMID 24349502.