Sulfolobus
| Sulfolobus | |
|---|---|
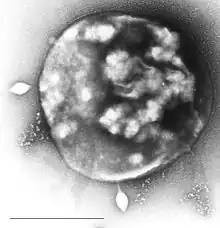 | |
| Electron micrograph of Sulfolobus infected with Sulfolobus virus STSV1. Bar = 1 μm. | |
| Scientific classification | |
| Domain: | |
| Kingdom: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | Sulfolobus Brock, Brock, Belly & Weiss 1972 |
| Type species | |
| Sulfolobus acidocaldarius Brock et al. 1972 | |
| Species | |
| |
Sulfolobus is a genus of microorganism in the family Sulfolobaceae. It belongs to the archaea domain.[2]
Sulfolobus species grow in volcanic springs with optimal growth occurring at pH 2-3 and temperatures of 75-80 °C, making them acidophiles and thermophiles respectively. Sulfolobus cells are irregularly shaped and flagellar.
Species of Sulfolobus are generally named after the location from which they were first isolated, e.g. Sulfolobus solfataricus was first isolated in the Solfatara volcano. Other species can be found throughout the world in areas of volcanic or geothermal activity, such as geological formations called mud pots, which are also known as solfatare (plural of solfatara).
Sulfolobus as a model to study the molecular mechanisms of DNA replication
When the first Archaeal genome, Methanococcus jannaschii, had been sequenced completely in 1996, it was found that the genes in the genome of Methanococcus jannaschii involved in DNA replication, transcription, and translation were more related to their counterparts in eukaryotes than to those in other prokaryotes. In 2001, the first genome sequence of Sulfolobus, Sulfolobus solfataricus P2, was published. In P2's genome, the genes related to chromosome replication were likewise found to be more related to those in eukaryotes. These genes include DNA polymerase, primase (including two subunits), MCM, CDC6/ORC1, RPA, RPC, and PCNA. In 2004, the origins of DNA replication of Sulfolobus solfataricus and Sulfolobus acidocaldarius were identified. It showed that both species contained two origins in their genome. This was the first time that more than a single origin of DNA replication had been shown to be used in a prokaryotic cell. The mechanism of DNA replication in archaea is evolutionary conserved, and similar to that of eukaryotes. Sulfolobus is now used as a model to study the molecular mechanisms of DNA replication in Archaea. And because the system of DNA replication in Archaea is much simpler than that in Eukaryota, it was suggested that Archaea could be used as a model to study the much more complex DNA replication in Eukaryota.
Role in biotechnology
Sulfolobus proteins are of interest for biotechnology and industrial use due to their thermostable nature. One application is the creation of artificial derivatives from S. acidocaldarius proteins, named affitins. Intracellular proteins are not necessarily stable at low pH however, as Sulfolobus species maintain a significant pH gradient across the outer membrane. Sulfolobales are metabolically dependent on sulfur: heterotrophic or autotrophic, their energy comes from the oxidation of sulfur and/or cellular respiration in which sulfur acts as the final electron acceptor. For example, S. tokodaii is known to oxidize hydrogen sulfide to sulfate intracellularly.
Phylogeny
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) [3] and National Center for Biotechnology Information (NCBI)[4]
| 16S rRNA-based LTP release 132 by The All-Species Living Tree Project[5] | Annotree v1.2.0[6][7] which uses the GTDB 05-RS95 (Genome Taxonomy Database)[8][9] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
Genome status
The complete genomes have been sequenced for S. acidocaldarius DSM 639 (2,225,959 nucleotides),[10] S. solfataricus P2 (2,992,245 nucleotides),[11] and S. tokodaii str. 7 (2,694,756 nucleotides).[12]
Genome structure
The archaeon Sulfolobus solfataricus has a circular chromosome that consists of 2,992,245 bp. Another sequenced species, S. tokodaii has a circular chromosome as well but is slightly smaller with 2,694,756 bp. Both species lack the genes ftsZ and minD, which has been characteristic of sequenced Crenarchaeota. They also code for citrate synthase and two subunits of 2-oxoacid:ferredoxin oxidoreductase, which plays the same role as alpha-ketoglutarate dehydrogenase in the TCA (tricarboxylic/Krebs/citric acid) cycle. This indicates that Sulfolobus has a TCA cycle system similar to that found in mitochondria of eukaryotes. Other genes in the respiratory chain which partake in the production of ATP were not similar to what is found in eukaryotes. Cytochrome c is one such example that plays an important role in electron transfer to oxygen in eukaryotes. This was also found in A. pernix K1. Since this step is important for an aerobic microorganism like Sulfolobus, it probably uses a different molecule for the same function or has a different pathway.
Cell structure and metabolism
Sulfolobus can grow either lithoautotrophically by oxidizing sulfur, or chemoheterotrophically using sulfur to oxidize simple reduced carbon compounds. Heterotrophic growth has only been observed, however, in the presence of oxygen. The principle metabolic pathways are a glycolytic pathway, a pentose phosphate pathway, and the TCA cycle.
All Archaea have lipids with ether links between the head group and side chains, making the lipids more resistant to heat and acidity than bacterial and eukaryotic ester-linked lipids. The Sulfolobales are known for unusual tetraether lipids. In Sulfolobales, the ether-linked lipids are joined covalently across the "bilayer," making tetraethers. Technically, therefore, the tetraethers form a monolayer, not a bilayer. The tetraethers help Sulfolobus species survive extreme acid as well as high temperature.
Ecology
S. solfataricus has been found in different areas including Yellowstone National Park, Mount St. Helens, Iceland, Italy, and Russia to name a few. Sulfolobus is located almost wherever there is volcanic activity. They thrive in environments where the temperature is about 80 °C with a pH at about 3 and sulfur present. Another species, S. tokodaii, has been located in an acidic spa in Beppu Hot Springs, Kyushu, Japan. Sediments from ~90m below the seafloor on the Peruvian continental margin are dominated by intact archaeal tetraethers, and a significant fraction of the community is sedimentary archaea taxonomically linked to the crenarchaeal Sulfolobales (Sturt, et al., 2004).
DNA damage response
Exposure of Sulfolobus solfataricus or Sulfolobus acidocaldarius to the DNA damaging agents UV-irradiation, bleomycin or mitomycin C induced cellular aggregation.[13][14] Other physical stressors, such as pH or temperature shift, did not induce aggregation, suggesting that induction of aggregation is caused specifically by DNA damage.[14] Ajon et al.[13] showed that UV-induced cellular aggregation mediates chromosomal marker exchange with high frequency in S. acidocaldarius. Recombination rates exceeded those of uninduced cultures by up to three orders of magnitude. Wood et al.[15] also showed that UV-irradiation increased the frequency of recombination due to genetic exchange in S. acidocaldarius. Frols et al.[14][16] and Ajon et al.[13] hypothesized that the UV-inducible DNA transfer process and subsequent homologous recombinational repair represents an important mechanism to maintain chromosome integrity in S. acidocaldarius and S. solfataricus. This response may be a primitive form of sexual interaction, similar to the more well-studied bacterial transformation that is also associated with DNA transfer between cells leading to homologous recombinational repair of DNA damage.[17][18]
The ups operon
The ups operon of Sulfolobus species is highly induced by UV irradiation. The pili encoded by this operon are employed in promoting cellular aggregation, which is necessary for subsequent DNA exchange between cells, resulting in homologous recombination. A study of the Sulfolobales acidocaldarius ups operon showed that one of the genes of the operon, saci-1497, encodes an endonuclease III that nicks UV-damaged DNA; and another gene of the operon, saci-1500, encodes a RecQ-like helicase that is able to unwind homologous recombination intermediates such as Holliday junctions.[19] It was proposed that Saci-1497 and Saci-1500 function in an homologous recombination-based DNA repair mechanism that uses transferred DNA as a template.[19] Thus it is thought that the ups system in combination with homologous recombination provide a DNA damage response which rescues Sulfolobales from DNA damaging threats.[19]
Sulfolobus as a viral host
Lysogenic viruses infect Sulfolobus for protection. The viruses cannot survive in the extremely acidic and hot conditions that Sulfolobus lives in, and so the viruses use Sulfolobus as protection against the harsh elements. This relationship allows the virus to replicate inside the archaea without being destroyed by the environment. The Sulfolobus viruses are temperate or permanent lysogens. Permanent lysogens differ from lysogenic bacteriophages in that the host cells are not lysed after the induction of Fuselloviridae production and eventually return to the lysogenic state. They are also unique in the sense that the genes encoding the structural proteins of the virus are constantly transcribed and DNA replication appears to be induced. The viruses infecting archaea like Sulfolobus have to use a strategy to escape prolonged direct exposure to the type of environment their host lives in, which may explain some of their unique properties.
See also
- Transformation (genetics)
- Evolution of sexual reproduction
References
- ↑ Dai, X; Wang, H; Zhang, Z; Li, K; Zhang, X; Mora-López, M; Jiang, C; Liu, C; Wang, L; Zhu, Y; Hernández-Ascencio, W; Dong, Z; Huang, L (2016). "Genome Sequencing of Sulfolobus sp. A20 from Costa Rica and Comparative Analyses of the Putative Pathways of Carbon, Nitrogen, and Sulfur Metabolism in Various Sulfolobus Strains". Frontiers in Microbiology. 7: 1902. doi:10.3389/fmicb.2016.01902. PMC 5127849. PMID 27965637.
- ↑ See the NCBI webpage on Sulfolobus. Data extracted from the "NCBI taxonomy resources". National Center for Biotechnology Information. Retrieved 2007-03-19.
- ↑ J.P. Euzéby. "Sulfolobus". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 2021-05-15.
- ↑ Sayers; et al. "Sulfolobus". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2021-05-15.
- ↑ All-Species Living Tree Project."16S rRNA-based LTP release 132". Silva Comprehensive Ribosomal RNA Database. Retrieved 2015-08-20.
- ↑ "AnnoTree v1.2.0". AnnoTree.
- ↑ Mendler, K; Chen, H; Parks, DH; Hug, LA; Doxey, AC (2019). "AnnoTree: visualization and exploration of a functionally annotated microbial tree of life". Nucleic Acids Research. 47 (9): 4442–4448. doi:10.1093/nar/gkz246. PMC 6511854. PMID 31081040.
- ↑ "GTDB release 05-RS95". Genome Taxonomy Database.
- ↑ Parks, DH; Chuvochina, M; Chaumeil, PA; Rinke, C; Mussig, AJ; Hugenholtz, P (September 2020). "A complete domain-to-species taxonomy for Bacteria and Archaea". Nature Biotechnology. 38 (9): 1079–1086. bioRxiv 10.1101/771964. doi:10.1038/s41587-020-0501-8. PMID 32341564. S2CID 216560589.
- ↑ Chen, L; Brügger, K; Skovgaard, M; Redder, P; She, Q; Torarinsson, E; Greve, B; Awayez, M; Zibat, A; Klenk, HP; Garrett, RA (July 2005). "The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota". Journal of Bacteriology. 187 (14): 4992–9. doi:10.1128/JB.187.14.4992-4999.2005. PMC 1169522. PMID 15995215.
- ↑ She, Q; Singh, RK; Confalonieri, F; Zivanovic, Y; Allard, G; Awayez, MJ; Chan-Weiher, CC; Clausen, IG; Curtis, BA; De Moors, A; Erauso, G; Fletcher, C; Gordon, PM; Heikamp-de Jong, I; Jeffries, AC; Kozera, CJ; Medina, N; Peng, X; Thi-Ngoc, HP; Redder, P; Schenk, ME; Theriault, C; Tolstrup, N; Charlebois, RL; Doolittle, WF; Duguet, M; Gaasterland, T; Garrett, RA; Ragan, MA; Sensen, CW; Van der Oost, J (3 July 2001). "The complete genome of the crenarchaeon Sulfolobus solfataricus P2". Proceedings of the National Academy of Sciences of the United States of America. 98 (14): 7835–40. Bibcode:2001PNAS...98.7835S. doi:10.1073/pnas.141222098. PMC 35428. PMID 11427726.
- ↑ Kawarabayasi, Y; Hino, Y; Horikawa, H; Jin-no, K; Takahashi, M; Sekine, M; Baba, S; Ankai, A; Kosugi, H; Hosoyama, A; Fukui, S; Nagai, Y; Nishijima, K; Otsuka, R; Nakazawa, H; Takamiya, M; Kato, Y; Yoshizawa, T; Tanaka, T; Kudoh, Y; Yamazaki, J; Kushida, N; Oguchi, A; Aoki, K; Masuda, S; Yanagii, M; Nishimura, M; Yamagishi, A; Oshima, T; Kikuchi, H (31 August 2001). "Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7". DNA Research. 8 (4): 123–40. doi:10.1093/dnares/8.4.123. PMID 11572479.
- 1 2 3 Ajon M; Fröls S; van Wolferen M; et al. (November 2011). "UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili" (PDF). Mol. Microbiol. 82 (4): 807–17. doi:10.1111/j.1365-2958.2011.07861.x. PMID 21999488.
- 1 2 3 Fröls S; Ajon M; Wagner M; et al. (November 2008). "UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation" (PDF). Mol. Microbiol. 70 (4): 938–52. doi:10.1111/j.1365-2958.2008.06459.x. PMID 18990182.
- ↑ Wood ER; Ghané F; Grogan DW (September 1997). "Genetic responses of the thermophilic archaeon Sulfolobus acidocaldarius to short-wavelength UV light". J. Bacteriol. 179 (18): 5693–8. doi:10.1128/jb.179.18.5693-5698.1997. PMC 179455. PMID 9294423.
- ↑ Fröls S; White MF; Schleper C (February 2009). "Reactions to UV damage in the model archaeon Sulfolobus solfataricus". Biochem. Soc. Trans. 37 (Pt 1): 36–41. doi:10.1042/BST0370036. PMID 19143598.
- ↑ Gross J; Bhattacharya D (2010). "Uniting sex and eukaryote origins in an emerging oxygenic world". Biol. Direct. 5: 53. doi:10.1186/1745-6150-5-53. PMC 2933680. PMID 20731852.
- ↑ Bernstein, H; Bernstein, C (2010). "Evolutionary Origin of Recombination during Meiosis". BioScience. 60 (7): 498–505. doi:10.1525/bio.2010.60.7.5. S2CID 86663600.
- 1 2 3 van Wolferen M, Ma X, Albers SV (2015). "DNA Processing Proteins Involved in the UV-Induced Stress Response of Sulfolobales". J. Bacteriol. 197 (18): 2941–51. doi:10.1128/JB.00344-15. PMC 4542170. PMID 26148716.
- Madigan M; Martinko J, eds. (2005). Brock Biology of Microorganisms (11th ed.). Prentice Hall. ISBN 978-0-13-144329-7.
Further reading
Scientific journals
- Judicial Commission of the International Committee on Systematics of Prokaryotes (2005). "The nomenclatural types of the orders Acholeplasmatales, Halanaerobiales, Halobacteriales, Methanobacteriales, Methanococcales, Methanomicrobiales, Planctomycetales, Prochlorales, Sulfolobales, Thermococcales, Thermoproteales and Verrucomicrobiales are the genera Acholeplasma, Halanaerobium, Halobacterium, Methanobacterium, Methanococcus, Methanomicrobium, Planctomyces, Prochloron, Sulfolobus, Thermococcus, Thermoproteus and Verrucomicrobium, respectively. Opinion 79". Int. J. Syst. Evol. Microbiol. 55 (Pt 1): 517–518. doi:10.1099/ijs.0.63548-0. PMID 15653928.
- Brock TD; Brock KM; Belly RT; Weiss RL (1972). "Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature". Arch. Mikrobiol. 84 (1): 54–68. doi:10.1007/BF00408082. PMID 4559703. S2CID 9204044.
Scientific books
- Stetter, KO (1989). "Order III. Sulfolobales ord. nov. Family Sulfolobaceae fam. nov.". In JT Staley; MP Bryant; N Pfennig; JG Holt (eds.). Bergey's Manual of Systematic Bacteriology. Vol. 3 (1st ed.). Baltimore: The Williams & Wilkins Co. p. 169.
Scientific databases
External links
- NCBI taxonomy page for Sulfolobus
- Search Tree of Life taxonomy pages for Sulfolobus
- Search Species2000 page for Sulfolobus
- MicrobeWiki page for Sulfolobus
- LPSN page for Sulfolobus
- Comparative Analysis of Sulfolobus Genomes (at DOE's IMG system)
- Sulfolobus Genome Projects (from Genomes OnLine Database)