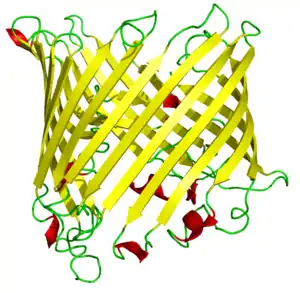
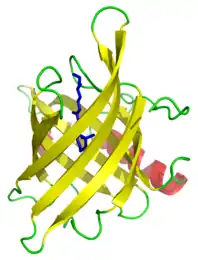
Beta barrel
In protein structures, a beta barrel is a beta sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand (hydrogen bond). Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and are commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria.[1] It has been shown that more than 600 proteins with various function (e.g., oxidase, dismutase, amylase) contain the beta barrel structure.[2]
In many cases, the strands contain alternating polar and non-polar (hydrophilic and hydrophobic) amino acids, so that the hydrophobic residues are oriented into the interior of the barrel to form a hydrophobic core and the polar residues are oriented toward the outside of the barrel on the solvent-exposed surface. Porins and other membrane proteins containing beta barrels reverse this pattern, with hydrophobic residues oriented toward the exterior where they contact the surrounding lipids, and hydrophilic residues oriented toward the aqueous interior pore.
All beta-barrels can be classified in terms of two integer parameters: the number of strands in the beta-sheet, n, and the "shear number", S, a measure of the stagger of the strands in the beta-sheet.[3] These two parameters (n and S) are related to the inclination angle of the beta strands relative to the axis of the barrel.[4][5][6]
Types
Up-and-down
Up-and-down barrels are the simplest barrel topology and consist of a series of beta strands, each of which is hydrogen-bonded to the strands immediately before and after it in the primary sequence.
Jelly roll
The jelly roll fold or barrel, also known as the Swiss roll, typically comprises eight beta strands arranged in two four-stranded sheets. Adjacent strands along the sequence alternate between the two sheets, such that they are "wrapped" in three dimensions to form a barrel shape.
Examples
Porins
Sixteen- or eighteen-stranded up-and-down beta barrel structures occur in porins, which function as transporters for ions and small molecules that cannot diffuse across a cellular membrane. Such structures appear in the outer membranes of gram-negative bacteria, chloroplasts, and mitochondria. The central pore of the protein, sometimes known as the eyelet, is lined with charged residues arranged so that the positive and negative charges appear on opposite sides of the pore. A long loop between two beta strands partially occludes the central channel; the exact size and conformation of the loop helps in discriminating between molecules passing through the transporter.
Preprotein translocases
Beta barrels also function within endosymbiont derived organelles such as mitochondria and chloroplasts to transport proteins.[8] Within the mitochondrion two complexes exist with beta barrels serving as the pore forming subunit, Tom40 of the Translocase of the outer membrane, and Sam50 of the Sorting and assembly machinery. The chloroplast also has functionally similar beta barrel containing complexes, the best characterised of which is Toc75 of the TOC complex (Translocon at the outer envelope membrane of chloroplasts).
Lipocalins
Lipocalins are typically eight-stranded up-and-down beta barrel proteins that are secreted into the extracellular environment. A distinctive feature is their ability to bind and transport small hydrophobic molecules in the barrel calyx. Examples of the family include retinol binding proteins (RBPs) and major urinary proteins (Mups). RBP binds and transports retinol (vitamin A), while Mups bind a number of small, organic pheromones, including 2-sec-butyl-4,5-dihydrothiazole (abbreviated as SBT or DHT), 6-hydroxy-6-methyl-3-heptanone (HMH) and 2,3 dihydro-exo-brevicomin (DHB).[9][10][11]
Shear number



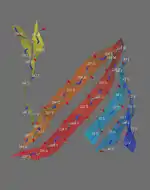
A piece of paper can be formed into a cylinder by bringing opposite sides together. The two edges come together to form a line. Shear can be created by sliding the two edges parallel to that line. Likewise, a beta barrel can be formed by bringing the edges of a beta sheet together to form a cylinder. If those edges are displaced, shear is created.
A similar definition is found in geology, where shear refers to a displacement within rock perpendicular to the rock surface. In physics, the amount of displacement is referred to as shear strain, which has units of length. For shear number in barrels, displacement is measured in units of amino acid residues.
The determination of shear number requires the assumption that each amino acid in one strand of a beta sheet is adjacent to just one amino acid in the neighboring strand (this assumption may not hold if, for example, a beta bulge is present).[12] To illustrate, S will be calculated for green fluorescent protein. This protein was chosen because the beta barrel contains both parallel and antiparallel strands. To determine which amino acid residues are adjacent in the beta strands, the location of hydrogen bonds is determined.
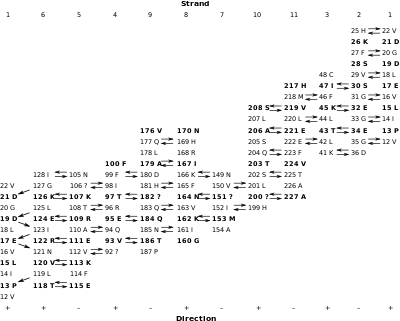
The inter-strand hydrogen bonds can be summarised in a table. Each column contains the residues in one strand (strand 1 is repeated in the last column). The arrows indicate the hydrogen bonds that were identified in the figures. The relative direction of each strand is indicated by the "+" and "-" at the bottom of the table. Except for strands 1 and 6, all strands are antiparallel. The parallel interaction between strands 1 and 6 accounts for the different appearance of the hydrogen bonding pattern. (Some arrows are missing because not all of the hydrogen bonds expected were identified. Non-standard amino acids are indicated with "?") The side chains that point to the outside of the barrel are in bold.
If no shear were present in this barrel, then residue 12 V, say, in strand 1 should end up in the last strand at the same level as it started at. However, because of shear, 12 V is not at the same level: it is 14 residues higher than it started at, so its shear number, S, is 14.
See also
- OMPdb (2011)
References
- Wimley WC (August 2003). "The versatile beta-barrel membrane protein". Current Opinion in Structural Biology. 13 (4): 404–411. doi:10.1016/S0959-440X(03)00099-X. PMID 12948769.
- Lu Y, Yeung N, Sieracki N, Marshall NM (August 2009). "Design of functional metalloproteins". Nature. 460 (7257): 855–862. Bibcode:2009Natur.460..855L. doi:10.1038/nature08304. PMC 2770889. PMID 19675646.
- Murzin AG, Lesk AM, Chothia C (March 1994). "Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis". Journal of Molecular Biology. 236 (5): 1369–1381. doi:10.1016/0022-2836(94)90064-7. PMID 8126726.
- Murzin AG, Lesk AM, Chothia C (March 1994). "Principles determining the structure of beta-sheet barrels in proteins. II. The observed structures". Journal of Molecular Biology. 236 (5): 1382–1400. doi:10.1016/0022-2836(94)90065-5. PMID 8126727.
- Liu WM (January 1998). "Shear numbers of protein beta-barrels: definition refinements and statistics". Journal of Molecular Biology. 275 (4): 541–545. doi:10.1006/jmbi.1997.1501. PMID 9466929.
- Hayward S, Milner-White EJ (October 2017). "Geometrical principles of homomeric β-barrels and β-helices: Application to modeling amyloid protofilaments" (PDF). Proteins. 85 (10): 1866–1881. doi:10.1002/prot.25341. PMID 28646497. S2CID 206410314.
- Böcskei Z, Groom CR, Flower DR, Wright CE, Phillips SE, Cavaggioni A, et al. (November 1992). "Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography". Nature. 360 (6400): 186–188. Bibcode:1992Natur.360..186B. doi:10.1038/360186a0. PMID 1279439. S2CID 4362015.
- Schleiff E, Soll J (November 2005). "Membrane protein insertion: mixing eukaryotic and prokaryotic concepts". EMBO Reports. 6 (11): 1023–1027. doi:10.1038/sj.embor.7400563. PMC 1371041. PMID 16264426.
- Halpern M, Martínez-Marcos A (June 2003). "Structure and function of the vomeronasal system: an update". Progress in Neurobiology. 70 (3): 245–318. doi:10.1016/S0301-0082(03)00103-5. PMID 12951145. S2CID 31122845.
- Timm DE, Baker LJ, Mueller H, Zidek L, Novotny MV (May 2001). "Structural basis of pheromone binding to mouse major urinary protein (MUP-I)". Protein Science. 10 (5): 997–1004. doi:10.1110/ps.52201. PMC 2374202. PMID 11316880.
- Armstrong SD, Robertson DH, Cheetham SA, Hurst JL, Beynon RJ (October 2005). "Structural and functional differences in isoforms of mouse major urinary proteins: a male-specific protein that preferentially binds a male pheromone". The Biochemical Journal. 391 (Pt 2): 343–350. doi:10.1042/BJ20050404. PMC 1276933. PMID 15934926.
- Nagano N, Hutchinson EG, Thornton JM (October 1999). "Barrel structures in proteins: automatic identification and classification including a sequence analysis of TIM barrels". Protein Science. 8 (10): 2072–2084. doi:10.1110/ps.8.10.2072. PMC 2144152. PMID 10548053.
Further reading
- Branden C, Tooze J (1999). Introduction to protein structure (2nd ed.). New York: Garland Pub. ISBN 978-0-8153-2304-4.
- Michalik M, Orwick-Rydmark M, Habeck M, Alva V, Arnold T, Linke D (2017). "An evolutionarily conserved glycine-tyrosine motif forms a folding core in outer membrane proteins". PLOS ONE. 12 (8): e0182016. Bibcode:2017PLoSO..1282016M. doi:10.1371/journal.pone.0182016. PMC 5542473. PMID 28771529.
- Hayward S, Milner-White EJ (October 2017). "Geometrical principles of homomeric β-barrels and β-helices: Application to modeling amyloid protofilaments" (PDF). Proteins. 85 (10): 1866–1881. doi:10.1002/prot.25341. PMID 28646497. S2CID 206410314.
External links
- Explanation of all-beta topologies: "orthogonal beta-sandwiches" are beta-barrels (as defined in this article); "aligned" beta-sandwiches" correspond to beta-sandwich folds in SCOP classification.
- all-beta folds in SCOP database (folds 54 to 100 are water-soluble beta-barrels).
- CATH database - folds and homologous superfamilies within the beta-barrel architecture.
- General classification and images of protein structures from Jane Richardson lab
- Images and examples of transmembrane beta-barrels
- Stockholm Bioinformatics Center review of transmembrane proteins
- The Lipocalin Website
- The OMPdb database for beta-barrel proteins