Argonaute
The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function,[1] plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi).[2] Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity (base pairing), which then leads to mRNA cleavage, translation inhibition, and/or the initiation of mRNA decay.[3]
The name of this protein family is derived from a mutant phenotype resulting from mutation of AGO1 in Arabidopsis thaliana, which was likened by Bohmert et al. to the appearance of the pelagic octopus Argonauta argo.[4]
| Argonaute Piwi domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 An argonaute protein from Pyrococcus furiosus. PDB 1U04. PIWI domain is on the right, PAZ domain to the left. | |||||||||
| Identifiers | |||||||||
| Symbol | Piwi | ||||||||
| Pfam | PF02171 | ||||||||
| InterPro | IPR003165 | ||||||||
| PROSITE | PS50822 | ||||||||
| CDD | cd02826 | ||||||||
| |||||||||
| Argonaute Paz domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Paz | ||||||||
| Pfam | PF12212 | ||||||||
| InterPro | IPR021103 | ||||||||
| SCOP2 | b.34.14.1 / SCOPe / SUPFAM | ||||||||
| |||||||||
RNA interference
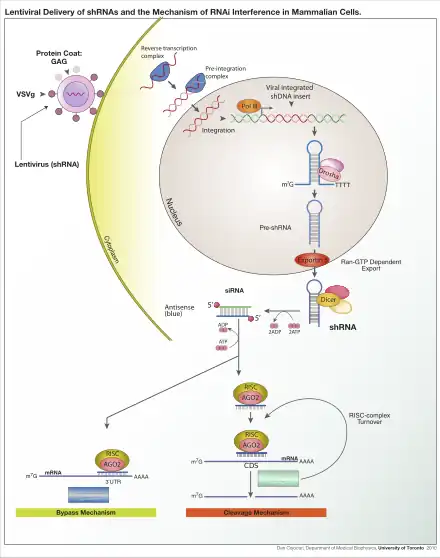
RNA interference (RNAi) is a biological process in which the RNA molecules inhibit gene expression. The method of inhibition is via the destruction of specific mRNA molecules or by simply suppressing the protein translation.[5] The RNA interference has a significant role in defending cells against parasitic nucleotide sequences. In many eukaryotes, including animals, the RNA interference pathway is found, and it is initiated by the enzyme Dicer. Dicer cleaves long double-stranded RNA (dsRNA, often found in viruses and small interfering RNA) molecules into short double stranded fragments of around 20 nucleotide siRNAs. The dsRNA is then separated into two single-stranded RNAs (ssRNA) – the passenger strand and the guide strand. Subsequently, the passenger strand is degraded, while the guide strand is incorporated into the RNA-induced silencing complex (RISC). The most well-studied outcome of the RNAi is post-transcriptional gene silencing, which occurs when the guide strand pairs with a complementary sequence in a messenger RNA molecule and induces cleavage by Argonaute, that lies in the core of RNA-induced silencing complex.
Argonaute proteins are the active part of RNA-induced silencing complex, cleaving the target mRNA strand complementary to their bound siRNA.[6] Theoretically the dicer produces short double-stranded fragments so there should be also two functional single-stranded siRNA produced. But only one of the two single-stranded RNA here will be utilized to base pair with target mRNA. It is known as the guide strand, incorporated into the Argonaute protein and leads gene silencing. The other single-stranded named passenger strand is degraded during the RNA-induced silencing complex process.[7]
Once the Argonaute is associated with the small RNA, the enzymatic activity conferred by the PIWI domain cleaves only the passenger strand of the small interfering RNA. RNA strand separation and incorporation into the Argonaute protein are guided by the strength of the hydrogen bond interaction at the 5′-ends of the RNA duplex, known as the asymmetry rule. Also the degree of complementarity between the two strands of the intermediate RNA duplex defines how the miRNA are sorted into different types of Argonaute proteins.
In animals, Argonaute associated with miRNA binds to the 3′-untranslated region of mRNA and prevents the production of proteins in various ways. The recruitment of Argonaute proteins to targeted mRNA can induce mRNA degradation. The Argonaute-miRNA complex can also affect the formation of functional ribosomes at the 5′-end of the mRNA. The complex here competes with the translation initiation factors and/or abrogate ribosome assembly. Also, the Argonaute-miRNA complex can adjust protein production by recruiting cellular factors such as peptides or post translational modifying enzymes, which degrade the growing of polypeptides.[8]
In plants, once de novo double-stranded (ds) RNA duplexes are generated with the target mRNA, an unknown RNase-III-like enzyme produces new siRNAs, which are then loaded onto the Argonaute proteins containing PIWI domains, lacking the catalytic amino acid residues, which might induce another level of specific gene silencing.
Functional domains and mechanism
The Argonaute (AGO) gene family encodes for six characteristic domains: N- terminal (N), Linker-1 (L1), PAZ, Linker-2 (L2), Mid, and a C-terminal PIWI domain.[8]
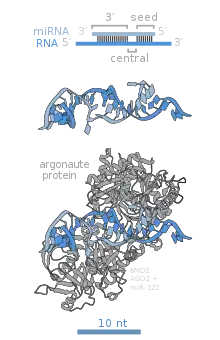
The name for the PAZ domain is an acronym made from the gene names of Drosophila piwi, Arabidopsis argonaute-1, and Arabidopsis zwille (also known as pinhead, and later renamed argonaute-10), where the domain was first recognized to be conserved. The PAZ domain is an RNA binding module that recognizes single-stranded 3′ ends of siRNA, miRNA and piRNA, in a sequence independent manner.
The Drosophila PIWI protein gave its name to this characteristic motif. Structurally resembling RNaseH, the PIWI domain is essential for the target cleavage. The active site with aspartate – aspartate – glutamate triad harbors a divalent metal ion, necessary for the catalysis. Family members of AGO that lost this conserved feature during evolution will lack the cleavage activity. In human AGO, the PIWI motif also mediates protein-protein interaction at the PIWI box, where it binds to Dicer at one of the RNase III domain.[9]
At the interface of PIWI and Mid domains sits the 5′ phosphate of a siRNA, miRNA or piRNA, which is found essential in the functionality. Within Mid lies a MC motif, a homologue structure proposed to mimic the cap-binding structure motif found in eIF4E. It was later found that the MC motif is not involved in mRNA cap binding [8]
Family members
In humans, there are eight AGO family members, some of which are investigated intensively. However, even though AGO1–4 are capable of loading miRNA, endonuclease activity and thus RNAi-dependent gene silencing exclusively belongs to AGO2. Considering the sequence conservation of PAZ and PIWI domains across the family, the uniqueness of AGO2 is presumed to arise from either the N-terminus or the spacing region linking PAZ and PIWI motifs.[9]
Several AGO family members in plants also attract study. AGO1 is involved in miRNA related RNA degradation, and plays a central role in morphogenesis. In some organisms, it is strictly required for epigenetic silencing. It is regulated by miRNA itself. AGO4 does not involve in RNAi directed RNA degradation, but in DNA methylation and other epigenetic regulation, through small RNA (smRNA) pathway. AGO10 is involved in plant development. AGO7 has a function distinct from AGO 1 and 10, and is not found in gene silencing induced by transgenes. Instead, it is related to developmental timing in plants.[10]
Disease and therapeutic tools
Argonaute proteins were reported to be associated with cancers.[11][12] For the diseases that are involved with selective or elevated expression of particular identified genes, such as pancreatic cancer, the high sequence specificity of RNA interference might make it suitable to be a suitable treatment, particularly appropriate for combating cancers associated with mutated endogenous gene sequences. It has been reported several tiny non-coding RNAs(microRNAs) are related with human cancers, like miR-15a and miR-16a are frequently deleted and/or down-regulated in patients. Even though the biological functions of miRNAs are not fully understood, the roles for miRNAs in the coordination of cell proliferation and cell death during development and metabolism have been uncovered. It is trusted that the miRNAs can direct negative or positive regulation at different levels, which depends on the specific miRNAs and target base pair interaction and the cofactors that recognize them.[13]
Because it has been widely known that many viruses have RNA rather than DNA as their genetic material and go through at least one stage in their life cycle when they make double-stranded RNA, RNA interference has been considered to be a potentially evolutionarily ancient mechanism for protecting organisms from viruses. The small interfering RNAs produced by Dicer cause sequence specific, post-transcriptional gene silencing by guiding an endonuclease, the RNA-induced silencing complex (RISC), to mRNA. This process has been seen in a wide range of organisms, such as Neurospora fungus (in which it is known as quelling), plants (post-transcriptional gene silencing) and mammalian cells(RNAi). If there is a complete or near complete sequence complementarity between the small RNA and the target, the Argonaute protein component of RISC mediates cleavage of the target transcript, the mechanism involves repression of translation predominantly.
Importantly, Argonaute 4 (AGO4)-deficient influenza-infected mice have significantly higher burden and viral titers in vivo[14] which is in contrast to AGO1 or AGO3-deficient mice.[15] So, specific promotion of AGO4 function in mammalian cells may be an effective antiviral strategy.
Biotechnological applications of prokaryotic Argonaute proteins
In 2016, a group from Hebei University of Science and Technology reported genome editing using a prokaryotic Argonaute protein from Natronobacterium gregoryi. However, evidence for application of Argonaute proteins as DNA-guided nucleases for genome editing have been questioned, with the retraction of the claim from the leading journal.[16] In 2017, a group from University of Illinois reported using a prokaryotic Argonaute protein taken from Pyrococcus furiosus (PfAgo) along with guide DNA to edit DNA in vitro as artificial restriction enzymes.[17] PfAgo based artificial restriction enzymes were also used for storing data on native DNA sequences via enzymatic nicking.[18]
References
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H (December 1998). "A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal". Genes & Development. 12 (23): 3715–3727. doi:10.1101/gad.12.23.3715. PMC 317255. PMID 9851978.
- Mauro M, Berretta M, Palermo G, Cavalieri V, La Rocca G (June 2022). "The multiplicity of Argonaute complexes in mammalian cells". J Pharmacol Exp Ther. doi:10.1124/jpet.122.001158. PMID 35667689.
- Jonas S, Izaurralde E (July 2015). "Towards a molecular understanding of microRNA-mediated gene silencing". Nature Reviews. Genetics. 16 (7): 421–433. doi:10.1038/nrg3965. PMID 26077373. S2CID 24892348.
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C (January 1998). "AGO1 defines a novel locus of Arabidopsis controlling leaf development". The EMBO Journal. 17 (1): 170–180. doi:10.1093/emboj/17.1.170. PMC 1170368. PMID 9427751.
- Guo H, Ingolia NT, Weissman JS, Bartel DP (August 2010). "Mammalian microRNAs predominantly act to decrease target mRNA levels". Nature. 466 (7308): 835–840. Bibcode:2010Natur.466..835G. doi:10.1038/nature09267. PMC 2990499. PMID 20703300.
- Kupferschmidt K (August 2013). "A lethal dose of RNA". Science. 341 (6147): 732–733. Bibcode:2013Sci...341..732K. doi:10.1126/science.341.6147.732. PMID 23950525.
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R (November 2005). "Human RISC couples microRNA biogenesis and posttranscriptional gene silencing". Cell. 123 (4): 631–640. doi:10.1016/j.cell.2005.10.022. PMID 16271387.
- Hutvagner G, Simard MJ (January 2008). "Argonaute proteins: key players in RNA silencing". Nature Reviews. Molecular Cell Biology. 9 (1): 22–32. doi:10.1038/nrm2321. hdl:10453/15429. PMID 18073770. S2CID 8822503.
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (July 2004). "Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs". Molecular Cell. 15 (2): 185–197. doi:10.1016/j.molcel.2004.07.007. PMID 15260970.
- Meins F, Si-Ammour A, Blevins T (2005). "RNA silencing systems and their relevance to plant development". Annual Review of Cell and Developmental Biology. 21 (1): 297–318. doi:10.1146/annurev.cellbio.21.122303.114706. PMID 16212497.
- Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H (June 2002). "Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas". Oncogene. 21 (25): 3988–3999. doi:10.1038/sj.onc.1205505. PMID 12037681. S2CID 6078065.
- Ross RJ, Weiner MM, Lin H (January 2014). "PIWI proteins and PIWI-interacting RNAs in the soma". Nature. 505 (7483): 353–359. doi:10.1038/nature12987. PMC 4265809. PMID 24429634.
- Hannon GJ (July 2002). "RNA interference". Nature. 418 (6894): 244–251. Bibcode:2002Natur.418..244H. doi:10.1038/418244a. PMID 12110901.
- Adiliaghdam F, Basavappa M, Saunders TL, Harjanto D, Prior JT, Cronkite DA, et al. (February 2020). "A Requirement for Argonaute 4 in Mammalian Antiviral Defense". Cell Reports. 30 (6): 1690–1701.e4. doi:10.1016/j.celrep.2020.01.021. PMC 7039342. PMID 32049003.
- Van Stry M, Oguin TH, Cheloufi S, Vogel P, Watanabe M, Pillai MR, et al. (April 2012). "Enhanced susceptibility of Ago1/3 double-null mice to influenza A virus infection". Journal of Virology. 86 (8): 4151–4157. doi:10.1128/JVI.05303-11. PMC 3318639. PMID 22318144.
- Cyranoski D (2017). "Authors retract controversial NgAgo gene-editing study". Nature. doi:10.1038/nature.2017.22412.
- Enghiad B, Zhao H (May 2017). "Programmable DNA-Guided Artificial Restriction Enzymes". ACS Synthetic Biology. 6 (5): 752–757. doi:10.1021/acssynbio.6b00324. PMID 28165224. S2CID 3833124.
- Tabatabaei SK, Wang B, Athreya NB, Enghiad B, Hernandez AG, Fields CJ, et al. (April 2020). "DNA punch cards for storing data on native DNA sequences via enzymatic nicking". Nature Communications. 11 (1): 1742. Bibcode:2020NatCo..11.1742T. doi:10.1038/s41467-020-15588-z. PMC 7142088. PMID 32269230.
External links
- starBase database: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq(HITS-CLIP, PAR-CLIP) and Degradome-Seq data.