Antifreeze protein
Antifreeze proteins (AFPs) or ice structuring proteins refer to a class of polypeptides produced by certain animals, plants, fungi and bacteria that permit their survival in temperatures below the freezing point of water. AFPs bind to small ice crystals to inhibit the growth and recrystallization of ice that would otherwise be fatal.[3] There is also increasing evidence that AFPs interact with mammalian cell membranes to protect them from cold damage. This work suggests the involvement of AFPs in cold acclimatization.[4]
| Insect antifreeze protein, Tenebrio-type | |||||||||
|---|---|---|---|---|---|---|---|---|---|
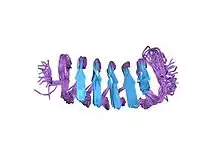 Structure of the Tenebrio molitor beta-helical antifreeze protein[1] | |||||||||
| Identifiers | |||||||||
| Symbol | AFP | ||||||||
| Pfam | PF02420 | ||||||||
| InterPro | IPR003460 | ||||||||
| SCOP2 | 1ezg / SCOPe / SUPFAM | ||||||||
| |||||||||
| Insect antifreeze protein (CfAFP) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
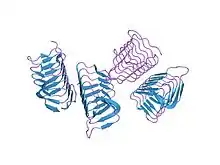 Structure of Choristoneura fumiferana (spruce budworm) beta-helical antifreeze protein[2] | |||||||||
| Identifiers | |||||||||
| Symbol | CfAFP | ||||||||
| Pfam | PF05264 | ||||||||
| InterPro | IPR007928 | ||||||||
| SCOP2 | 1m8n / SCOPe / SUPFAM | ||||||||
| |||||||||
| Fish antifreeze protein, type I | |
|---|---|
| Identifiers | |
| Symbol | ? |
| InterPro | IPR000104 |
| SCOP2 | 1wfb / SCOPe / SUPFAM |
| Fish antifreeze protein, type II | |
|---|---|
| Identifiers | |
| Symbol | ? |
| InterPro | IPR002353 |
| CATH | 2py2 |
| SCOP2 | 2afp / SCOPe / SUPFAM |
| Fish antifreeze protein, type III | |
|---|---|
| Identifiers | |
| Symbol | ? |
| InterPro | IPR006013 |
| SCOP2 | 1hg7 / SCOPe / SUPFAM |
| See also the SAF domain (InterPro: IPR013974). | |
| Ice-binding protein-like (sea ice organism) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | DUF3494 | ||||||||
| Pfam | PF11999 | ||||||||
| InterPro | IPR021884 | ||||||||
| |||||||||
Non-colligative properties
Unlike the widely used automotive antifreeze, ethylene glycol, AFPs do not lower freezing point in proportion to concentration. Rather, they work in a noncolligative manner. This phenomenon allows them to act as an antifreeze at concentrations 1/300th to 1/500th of those of other dissolved solutes. Their low concentration minimizes their effect on osmotic pressure.[4] The unusual properties of AFPs are attributed to their selective affinity for specific crystalline ice forms and the resulting blockade of the ice-nucleation process.[5]
Thermal hysteresis
AFPs create a difference between the melting point and freezing point (busting temperature of AFP bound ice crystal) known as thermal hysteresis. The addition of AFPs at the interface between solid ice and liquid water inhibits the thermodynamically favored growth of the ice crystal. Ice growth is kinetically inhibited by the AFPs covering the water-accessible surfaces of ice.[5]
Thermal hysteresis is easily measured in the lab with a nanolitre osmometer. Organisms differ in their values of thermal hysteresis. The maximum level of thermal hysteresis shown by fish AFP is approximately −3.5 °C (Sheikh Mahatabuddin et al., SciRep)(29.3 °F). In contrast, aquatic organisms are exposed only to −1 to −2 °C below freezing. During the extreme winter months, the spruce budworm resists freezing at temperatures approaching −30 °C.[4]
The rate of cooling can influence the thermal hysteresis value of AFPs. Rapid cooling can substantially decrease the nonequilibrium freezing point, and hence the thermal hysteresis value. Consequently, organisms cannot necessarily adapt to their subzero environment if the temperature drops abruptly.[4]
Freeze tolerance versus freeze avoidance
Species containing AFPs may be classified as
Freeze avoidant: These species are able to prevent their body fluids from freezing altogether. Generally, the AFP function may be overcome at extremely cold temperatures, leading to rapid ice growth and death.
Freeze tolerant: These species are able to survive body fluid freezing. Some freeze tolerant species are thought to use AFPs as cryoprotectants to prevent the damage of freezing, but not freezing altogether. The exact mechanism is still unknown. However, it is thought AFPs may inhibit recrystallization and stabilize cell membranes to prevent damage by ice.[6] They may work in conjunction with ice nucleating proteins (INPs) to control the rate of ice propagation following freezing.[6]
Diversity
There are many known nonhomologous types of AFPs.
Fish AFPs

Antifreeze glycoproteins or AFGPs are found in Antarctic notothenioids and northern cod. They are 2.6-3.3 kD.[7] AFGPs evolved separately in notothenioids and northern cod. In notothenioids, the AFGP gene arose from an ancestral trypsinogen-like serine protease gene.[8]
- Type I AFP is found in winter flounder, longhorn sculpin and shorthorn sculpin. It is the best documented AFP because it was the first to have its three-dimensional structure determined.[9] Type I AFP consists of a single, long, amphipathic alpha helix, about 3.3-4.5 kD in size. There are three faces to the 3D structure: the hydrophobic, hydrophilic, and Thr-Asx face.[9]
- Type I-hyp AFP (where hyp stands for hyperactive) are found in several righteye flounders. It is approximately 32 kD (two 17 kD dimeric molecules). The protein was isolated from the blood plasma of winter flounder. It is considerably better at depressing freezing temperature than most fish AFPs.[10] The ability is partially derived from its many repeats of the Type I ice-binding site.[11]
- Type II AFPs (e.g. P05140) are found in sea raven, smelt and herring. They are cysteine-rich globular proteins containing five disulfide bonds.[12] Type II AFPs likely evolved from calcium dependent (c-type) lectins.[13] Sea ravens, smelt, and herring are quite divergent lineages of teleost. If the AFP gene were present in the most recent common ancestor of these lineages, it is peculiar that the gene is scattered throughout those lineages, present in some orders and absent in others. It has been suggested that lateral gene transfer could be attributed to this discrepancy, such that the smelt acquired the type II AFP gene from the herring.[14]
- Type III AFPs are found in Antarctic eelpout. They exhibit similar overall hydrophobicity at ice binding surfaces to type I AFPs. They are approximately 6kD in size.[7] Type III AFPs likely evolved from a sialic acid synthase (SAS) gene present in Antarctic eelpout. Through a gene duplication event, this gene—which has been shown to exhibit some ice-binding activity of its own—evolved into an effective AFP gene by loss of the N-terminal part.[15]
- Type IV AFPs (P80961) are found in longhorn sculpins. They are alpha helical proteins rich in glutamate and glutamine.[16] This protein is approximately 12KDa in size and consists of a 4-helix bundle.[16] Its only posttranslational modification is a pyroglutamate residue, a cyclized glutamine residue at its N-terminus.[16]
Plant AFPs
The classification of AFPs became more complicated when antifreeze proteins from plants were discovered.[17] Plant AFPs are rather different from the other AFPs in the following aspects:
- They have much weaker thermal hysteresis activity when compared to other AFPs.[18]
- Their physiological function is likely in inhibiting the recrystallization of ice rather than in preventing ice formation.[18]
- Most of them are evolved pathogenesis-related proteins, sometimes retaining antifungal properties.[18]
Insect AFPs
There are a number of AFPs found in insects, including those from Dendroides, Tenebrio and Rhagium beetles, spruce budworm and pale beauty moths, and midges (same order as flies). Insect AFPs share certain similarities, with most having higher activity (i.e. greater thermal hysteresis value, termed hyperactive) and a repetitive structure with a flat ice-binding surface. Those from the closely related Tenebrio and Dendroides beetles are homologous and each 12–13 amino-acid repeat is stabilized by an internal disulfide bond. Isoforms have between 6 and 10 of these repeats that form a coil, or beta-solenoid. One side of the solenoid has a flat ice-binding surface that consists of a double row of threonine residues.[6][19] Other beetles (genus Rhagium) have longer repeats without internal disulfide bonds that form a compressed beta-solenoid (beta sandwich) with four rows of threonine residus,[20] and this AFP is structurally similar to that modelled for the non-homologous AFP from the pale beauty moth.[21] In contrast, the AFP from the spruce budworm moth is a solenoid that superficially resembles the Tenebrio protein, with a similar ice-binding surface, but it has a triangular cross-section, with longer repeats that lack the internal disulfide bonds. The AFP from midges is structurally similar to those from Tenebrio and Dendroides, but the disulfide-braced beta-solenoid is formed from shorter 10 amino-acids repeats, and instead of threonine, the ice-binding surface consists of a single row of tyrosine residues.[22] Springtails (Collembola) are not insects, but like insects, they are arthropods with six legs. A species found in Canada, which is often called a "snow flea", produces hyperactive AFPs.[23] Although they are also repetitive and have a flat ice-binding surface, the similarity ends there. Around 50% of the residues are glycine (Gly), with repeats of Gly-Gly- X or Gly-X-X, where X is any amino acid. Each 3-amino-acid repeat forms one turn of a polyproline type II helix. The helices then fold together, to form a bundle that is two helices thick, with an ice-binding face dominated by small hydrophobic residues like alanine, rather than threonine.[24] Other insects, such as an Alaskan beetle, produce hyperactive antifreezes that are even less similar, as they are polymers of sugars (xylomannan) rather than polymers of amino acids (proteins).[25] Taken together, this suggests that most of the AFPs and antifreezes arose after the lineages that gave rise to these various insects diverged. The similarities they do share are the result of convergent evolution.
Sea ice organism AFPs
Many microorganisms living in sea ice possess AFPs that belong to a single family. The diatoms Fragilariopsis cylindrus and F. curta play a key role in polar sea ice communities, dominating the assemblages of both platelet layer and within pack ice. AFPs are widespread in these species, and the presence of AFP genes as a multigene family indicates the importance of this group for the genus Fragilariopsis.[26] AFPs identified in F. cylindrus belong to an AFP family which is represented in different taxa and can be found in other organisms related to sea ice (Colwellia spp., Navicula glaciei, Chaetoceros neogracile and Stephos longipes and Leucosporidium antarcticum)[27][28] and Antarctic inland ice bacteria (Flavobacteriaceae),[29][30] as well as in cold-tolerant fungi (Typhula ishikariensis, Lentinula edodes and Flammulina populicola).[31][32]
Several structures for sea ice AFPs have been solved. This family of proteins fold into a beta helix that form a flat ice-binding surface.[33] Unlike the other AFPs, there is not a singular sequence motif for the ice-binding site.[34]
AFP found from the metagenome of the ciliate Euplotes focardii and psychrophilic bacteria has an efficient ice re-crystallization inhibition ability.[35] 1 μM of Euplotes focardii consortium ice-binding protein (EfcIBP) is enough for the total inhibition of ice re-crystallization in –7.4 °C temperature. This ice-recrystallization inhibition ability helps bacteria to tolerate ice rather than preventing the formation of ice. EfcIBP produces also thermal hysteresis gap, but this ability is not as efficient as the ice-recrystallization inhibition ability. EfcIBP helps to protect both purified proteins and whole bacterial cells in freezing temperatures. Green fluorescent protein is functional after several cycles of freezing and melting when incubated with EfcIBP. Escherichia coli survives longer periods in 0 °C temperature when the efcIBP gene was inserted to E. coli genome.[35] EfcIBP has a typical AFP structure consisting of multiple beta-sheets and an alpha-helix. Also, all the ice-binding polar residues are at the same site of the protein.[35]
Evolution
The remarkable diversity and distribution of AFPs suggest the different types evolved recently in response to sea level glaciation occurring 1–2 million years ago in the Northern hemisphere and 10-30 million years ago in Antarctica. Data collected from deep sea ocean drilling has revealed that the development of the Antarctic Circumpolar Current was formed over 30 million years ago.[36] The cooling of Antarctic imposed from this current caused a mass extinction of teleost species that were unable to withstand freezing temperatures.[37] Notothenioids species with the antifreeze gylcoprotein were able to survive the glaciation event and diversify into new niches.[37][8]
This independent development of similar adaptations is referred to as convergent evolution.[4] Evidence for convergent evolution in Northern cod (Gadidae) and Notothenioids is supported by the findings of different spacer sequences and different organization of introns and exons as well as unmatching AFGP tripeptide sequences, which emerged from duplications of short ancestral sequences which were differently permuted (for the same tripeptide) by each group. These groups diverged approximately 7-15 million years ago. Shortly after (5-15 mya), the AFGP gene evolved from an ancestral pancreatic trypsinogen gene in Notothenioids. AFGP and trypsinogen genes split via a sequence divergence - an adaptation which occurred alongside the cooling and eventual freezing of the Antarctic Ocean. The evolution of the AFGP gene in Northern cod occurred more recently (~3.2 mya) and emerged from a noncoding sequence via tandem duplications in a Thr-Ala-Ala unit. Antarctic notothenioid fish and artic cod, Boreogadus saida, are part of two distinct orders and have very similar antifreeze glycoproteins.[38] Although the two fish orders have similar antifreeze proteins, cod species contain arginine in AFG, while Antarctic notothenioid do not.[38] The role of arginine as an enhancer has been investigated in Dendroides canadensis antifreeze protein (DAFP-1) by observing the effect of a chemical modification using 1-2 cyclohexanedione.[39] Previous research has found various enhancers of this bettles' antifreeze protein including a thaumatin-like protein and polycarboxylates.[40][41] Modifications of DAFP-1 with the arginine specific reagent resulted in the partial and complete loss of thermal hysteresis in DAFP-1, indicating that arginine plays a crucial role in enhancing its ability.[39] Different enhancer molecules of DAFP-1 have distinct thermal hysteresis activity.[41] Amornwittawat et al. 2008 found that the number of carboxylate groups in a molecules influence the enhancing ability of DAFP-1.[41] Optimum activity in TH is correlated with high concentration of enhancer molecules.[41] Li et al. 1998 investigated the effects of pH and solute on thermal hysteresis in Antifreeze proteins from Dendrioides canadensis.[42] TH activity of DAFP-4 was not affected by pH unless the there was a low solute concentration (pH 1) in which TH decreased.[42] The effect of five solutes; succinate, citrate, malate, malonate, and acetate, on TH activity was reported.[42] Among the five solutes, citrate was shown to have the greatest enhancing effect.[42]
This is an example of a proto-ORF model, a rare occurrence where new genes pre exist as a formed open reading frame before the existence of the regulatory element needed to activate them.
In fishes, horizontal gene transfer is responsible for the presence of Type II AFP proteins in some groups without a recently shared phylogeny. In Herring and smelt, up to 98% of introns for this gene are shared; the method of transfer is assumed to occur during mating via sperm cells exposed to foreign DNA.[43] The direction of transfer is known to be from herring to smelt as herring have 8 times the copies of AFP gene as smelt (1) and the segments of the gene in smelt house transposable elements which are otherwise characteristic of and common in herring but not found in other fishes.[43]
There are two reasons why many types of AFPs are able to carry out the same function despite their diversity:
- Although ice is uniformly composed of water molecules, it has many different surfaces exposed for binding. Different types of AFPs may interact with different surfaces.
- Although the five types of AFPs differ in their primary structure of amino acids, when each folds into a functioning protein they may share similarities in their three-dimensional or tertiary structure that facilitates the same interactions with ice.[4][44]
Antifreeze glycoprotein activity has been observed across several ray-finned species including eelpouts, sculpins, and cod species.[45][46] Fish species that possess the antifreeze glycoprotein express different levels of protein activity.[47] Polar cod (Boreogadus saida) exhibit similar protein activity and properties to the Antarctic species, T. borchgrevinki.[47] Both species have higher protein activity than saffron cod (Eleginus gracilis).[47] Ice antifreeze proteins have been reported in diatom species to help decrease the freezing point of organism's proteins.[26] Bayer-Giraldi et al. 2010 found 30 species from distinct taxa with homologues of ice antifreeze proteins.[26] The diversity is consistent with previous research that has observed the presence of these genes in crustaceans, insects, bacteria, and fungi.[8][48][49] Horizontal gene transfer is responsible for the presence of ice antifreeze proteins in two sea diatom species, F. cylindrus and F. curta.[26]
Mechanisms of action
AFPs are thought to inhibit ice growth by an adsorption–inhibition mechanism.[50] They adsorb to nonbasal planes of ice, inhibiting thermodynamically-favored ice growth.[51] The presence of a flat, rigid surface in some AFPs seems to facilitate its interaction with ice via Van der Waals force surface complementarity.[52]
Binding to ice
Normally, ice crystals grown in solution only exhibit the basal (0001) and prism faces (1010), and appear as round and flat discs.[5] However, it appears the presence of AFPs exposes other faces. It now appears the ice surface 2021 is the preferred binding surface, at least for AFP type I.[53] Through studies on type I AFP, ice and AFP were initially thought to interact through hydrogen bonding (Raymond and DeVries, 1977). However, when parts of the protein thought to facilitate this hydrogen bonding were mutated, the hypothesized decrease in antifreeze activity was not observed. Recent data suggest hydrophobic interactions could be the main contributor.[54] It is difficult to discern the exact mechanism of binding because of the complex water-ice interface. Currently, attempts to uncover the precise mechanism are being made through use of molecular modelling programs (molecular dynamics or the Monte Carlo method).[3][5]
Binding mechanism and antifreeze function
According to the structure and function study on the antifreeze protein from Pseudopleuronectes americanus,[55] the antifreeze mechanism of the type-I AFP molecule was shown to be due to the binding to an ice nucleation structure in a zipper-like fashion through hydrogen bonding of the hydroxyl groups of its four Thr residues to the oxygens along the direction in ice lattice, subsequently stopping or retarding the growth of ice pyramidal planes so as to depress the freeze point.[55]
The above mechanism can be used to elucidate the structure-function relationship of other antifreeze proteins with the following two common features:
History
In the 1950s, Norwegian scientist Scholander set out to explain how Arctic fish can survive in water colder than the freezing point of their blood. His experiments led him to believe there was “antifreeze” in the blood of Arctic fish.[3] Then in the late 1960s, animal biologist Arthur DeVries was able to isolate the antifreeze protein through his investigation of Antarctic fish.[56] These proteins were later called antifreeze glycoproteins (AFGPs) or antifreeze glycopeptides to distinguish them from newly discovered nonglycoprotein biological antifreeze agents (AFPs). DeVries worked with Robert Feeney (1970) to characterize the chemical and physical properties of antifreeze proteins.[57] In 1992, Griffith et al. documented their discovery of AFP in winter rye leaves.[17] Around the same time, Urrutia, Duman and Knight (1992) documented thermal hysteresis protein in angiosperms.[58] The next year, Duman and Olsen noted AFPs had also been discovered in over 23 species of angiosperms, including ones eaten by humans.[59] They reported their presence in fungi and bacteria as well.
Name change
Recent attempts have been made to relabel antifreeze proteins as ice structuring proteins to more accurately represent their function and to dispose of any assumed negative relation between AFPs and automotive antifreeze, ethylene glycol. These two things are completely separate entities, and show loose similarity only in their function.[60]
Commercial and medical applications
Numerous fields would be able to benefit from the protection of tissue damage by freezing. Businesses are currently investigating the use of these proteins in:
- Increasing freeze tolerance of crop plants and extending the harvest season in cooler climates
- Improving farm fish production in cooler climates
- Lengthening shelf life of frozen foods
- Improving cryosurgery
- Enhancing preservation of tissues for transplant or transfusion in medicine[23]
- Therapy for hypothermia
- Human Cryopreservation (Cryonics)
Unilever has obtained UK, US, EU, Mexico, China, Philippines, Australia and New Zealand approval to use a genetically modified yeast to produce antifreeze proteins from fish for use in ice cream production.[61][62] They are labeled "ISP" or ice structuring protein on the label, instead of AFP or antifreeze protein.
Recent news
One recent, successful business endeavor has been the introduction of AFPs into ice cream and yogurt products. This ingredient, labelled ice-structuring protein, has been approved by the Food and Drug Administration. The proteins are isolated from fish and replicated, on a larger scale, in genetically modified yeast.[63]
There is concern from organizations opposed to genetically modified organisms (GMOs) who believe that antifreeze proteins may cause inflammation.[64] Intake of AFPs in diet is likely substantial in most northerly and temperate regions already.[7] Given the known historic consumption of AFPs, it is safe to conclude their functional properties do not impart any toxicologic or allergenic effects in humans.[7]
As well, the transgenic process of ice structuring proteins production is widely used in society. Insulin and rennet are produced using this technology. The process does not impact the product; it merely makes production more efficient and prevents the death of fish that would otherwise be killed to extract the protein.
Currently, Unilever incorporates AFPs into some of its American products, including some Popsicle ice pops and a new line of Breyers Light Double Churned ice cream bars. In ice cream, AFPs allow the production of very creamy, dense, reduced fat ice cream with fewer additives.[65] They control ice crystal growth brought on by thawing on the loading dock or kitchen table, which reduces texture quality.[66]
In November 2009, the Proceedings of the National Academy of Sciences published the discovery of a molecule in an Alaskan beetle that behaves like AFPs, but is composed of saccharides and fatty acids.[25]
A 2010 study demonstrated the stability of superheated water ice crystals in an AFP solution, showing that while the proteins can inhibit freezing, they can also inhibit melting.[67] In 2021, EPFL and Warwick scientists have found an artificial imitation of antifreeze proteins.[68]
References
- Daley ME, Spyracopoulos L, Jia Z, Davies PL, Sykes BD (April 2002). "Structure and dynamics of a beta-helical antifreeze protein". Biochemistry. 41 (17): 5515–25. doi:10.1021/bi0121252. PMID 11969412.
- Leinala EK, Davies PL, Doucet D, Tyshenko MG, Walker VK, Jia Z (September 2002). "A beta-helical antifreeze protein isoform with increased activity. Structural and functional insights". The Journal of Biological Chemistry. 277 (36): 33349–52. doi:10.1074/jbc.M205575200. PMID 12105229.
- Goodsell D (December 2009). "Molecule of the Month: Antifreeze Proteins". The Scripps Research Institute and the RCSB PDB. doi:10.2210/rcsb_pdb/mom_2009_12. Archived from the original on 2015-11-04. Retrieved 2012-12-30.
- Fletcher GL, Hew CL, Davies PL (2001). "Antifreeze proteins of teleost fishes". Annual Review of Physiology. 63: 359–90. doi:10.1146/annurev.physiol.63.1.359. PMID 11181960.
- Jorov A, Zhorov BS, Yang DS (June 2004). "Theoretical study of interaction of winter flounder antifreeze protein with ice". Protein Science. 13 (6): 1524–37. doi:10.1110/ps.04641104. PMC 2279984. PMID 15152087.
- Duman JG (2001). "Antifreeze and ice nucleator proteins in terrestrial arthropods". Annual Review of Physiology. 63: 327–57. doi:10.1146/annurev.physiol.63.1.327. PMID 11181959.
- Crevel RW, Fedyk JK, Spurgeon MJ (July 2002). "Antifreeze proteins: characteristics, occurrence and human exposure". Food and Chemical Toxicology. 40 (7): 899–903. doi:10.1016/S0278-6915(02)00042-X. PMID 12065210.
- Chen L, DeVries AL, Cheng CH (April 1997). "Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish". Proceedings of the National Academy of Sciences of the United States of America. 94 (8): 3811–6. Bibcode:1997PNAS...94.3811C. doi:10.1073/pnas.94.8.3811. PMC 20523. PMID 9108060.
- Duman JG, de Vries AL (1976). "Isolation, characterization, and physical properties of protein antifreezes from the winter flounder, Pseudopleuronectes americanus". Comparative Biochemistry and Physiology. B, Comparative Biochemistry. 54 (3): 375–80. doi:10.1016/0305-0491(76)90260-1. PMID 1277804.
- Scotter AJ, Marshall CB, Graham LA, Gilbert JA, Garnham CP, Davies PL (October 2006). "The basis for hyperactivity of antifreeze proteins". Cryobiology. 53 (2): 229–39. doi:10.1016/j.cryobiol.2006.06.006. PMID 16887111.
- Graham LA, Marshall CB, Lin FH, Campbell RL, Davies PL (February 2008). "Hyperactive antifreeze protein from fish contains multiple ice-binding sites". Biochemistry. 47 (7): 2051–63. doi:10.1021/bi7020316. PMID 18225917.
- Ng NF, Hew CL (August 1992). "Structure of an antifreeze polypeptide from the sea raven. Disulfide bonds and similarity to lectin-binding proteins". The Journal of Biological Chemistry. 267 (23): 16069–75. doi:10.1016/S0021-9258(18)41967-9. PMID 1644794.
- Ewart KV, Rubinsky B, Fletcher GL (May 1992). "Structural and functional similarity between fish antifreeze proteins and calcium-dependent lectins". Biochemical and Biophysical Research Communications. 185 (1): 335–40. doi:10.1016/s0006-291x(05)90005-3. PMID 1599470.
- Graham LA, Lougheed SC, Ewart KV, Davies PL (July 2008). "Lateral transfer of a lectin-like antifreeze protein gene in fishes". PLOS ONE. 3 (7): e2616. Bibcode:2008PLoSO...3.2616G. doi:10.1371/journal.pone.0002616. PMC 2440524. PMID 18612417.

- Kelley JL, Aagaard JE, MacCoss MJ, Swanson WJ (August 2010). "Functional diversification and evolution of antifreeze proteins in the antarctic fish Lycodichthys dearborni". Journal of Molecular Evolution. 71 (2): 111–8. Bibcode:2010JMolE..71..111K. doi:10.1007/s00239-010-9367-6. PMID 20686757. S2CID 25737518.
- Deng G, Andrews DW, Laursen RA (January 1997). "Amino acid sequence of a new type of antifreeze protein, from the longhorn sculpin Myoxocephalus octodecimspinosis". FEBS Letters. 402 (1): 17–20. doi:10.1016/S0014-5793(96)01466-4. PMID 9013849.
- Griffith M, Ala P, Yang DS, Hon WC, Moffatt BA (October 1992). "Antifreeze protein produced endogenously in winter rye leaves". Plant Physiology. 100 (2): 593–6. doi:10.1104/pp.100.2.593. PMC 1075599. PMID 16653033.
- Griffith M, Yaish MW (August 2004). "Antifreeze proteins in overwintering plants: a tale of two activities". Trends in Plant Science. 9 (8): 399–405. doi:10.1016/j.tplants.2004.06.007. PMID 15358271.
- Liou YC, Tocilj A, Davies PL, Jia Z (July 2000). "Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein". Nature. 406 (6793): 322–4. Bibcode:2000Natur.406..322L. doi:10.1038/35018604. PMID 10917536. S2CID 4385352.
- Hakim A, Nguyen JB, Basu K, Zhu DF, Thakral D, Davies PL, et al. (April 2013). "Crystal structure of an insect antifreeze protein and its implications for ice binding". The Journal of Biological Chemistry. 288 (17): 12295–304. doi:10.1074/jbc.M113.450973. PMC 3636913. PMID 23486477.
- Lin FH, Davies PL, Graham LA (May 2011). "The Thr- and Ala-rich hyperactive antifreeze protein from inchworm folds as a flat silk-like β-helix". Biochemistry. 50 (21): 4467–78. doi:10.1021/bi2003108. PMID 21486083.
- Basu K, Wasserman SS, Jeronimo PS, Graham LA, Davies PL (April 2016). "Intermediate activity of midge antifreeze protein is due to a tyrosine-rich ice-binding site and atypical ice plane affinity". The FEBS Journal. 283 (8): 1504–15. doi:10.1111/febs.13687. PMID 26896764. S2CID 37207016.
- Graham LA, Davies PL (October 2005). "Glycine-rich antifreeze proteins from snow fleas". Science. 310 (5747): 461. doi:10.1126/science.1115145. PMID 16239469. S2CID 30032276.*Lay summary in: "New Antifreeze Protein Found In Fleas May Allow Longer Storage Of Transplant Organs". ScienceDaily. October 21, 2005.
- Pentelute BL, Gates ZP, Tereshko V, Dashnau JL, Vanderkooi JM, Kossiakoff AA, Kent SB (July 2008). "X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers". Journal of the American Chemical Society. 130 (30): 9695–701. doi:10.1021/ja8013538. PMC 2719301. PMID 18598029.
- Walters KR, Serianni AS, Sformo T, Barnes BM, Duman JG (December 2009). "A nonprotein thermal hysteresis-producing xylomannan antifreeze in the freeze-tolerant Alaskan beetle Upis ceramboides". Proceedings of the National Academy of Sciences of the United States of America. 106 (48): 20210–5. Bibcode:2009PNAS..10620210W. doi:10.1073/pnas.0909872106. PMC 2787118. PMID 19934038. S2CID 25741145.
- Bayer-Giraldi M, Uhlig C, John U, Mock T, Valentin K (April 2010). "Antifreeze proteins in polar sea ice diatoms: diversity and gene expression in the genus Fragilariopsis". Environmental Microbiology. 12 (4): 1041–52. doi:10.1111/j.1462-2920.2009.02149.x. PMID 20105220.
- Raymond JA, Fritsen C, Shen K (August 2007). "An ice-binding protein from an Antarctic sea ice bacterium". FEMS Microbiology Ecology. 61 (2): 214–21. doi:10.1111/j.1574-6941.2007.00345.x. PMID 17651136.
- Kiko R (April 2010). "Acquisition of freeze protection in a sea-ice crustacean through horizontal gene transfer?". Polar Biology. 33 (4): 543–56. doi:10.1007/s00300-009-0732-0. S2CID 20952951.
- Raymond JA, Christner BC, Schuster SC (September 2008). "A bacterial ice-binding protein from the Vostok ice core". Extremophiles. 12 (5): 713–7. doi:10.1007/s00792-008-0178-2. PMID 18622572. S2CID 505953.
- Xiao N, Inaba S, Tojo M, Degawa Y, Fujiu S, Kudoh S, Hoshino T (2010-12-22). "Antifreeze activities of various fungi and Stramenopila isolated from Antarctica". North American Fungi. 5: 215–220. doi:10.2509/naf2010.005.00514.
- Hoshino T, Kiriaki M, Ohgiya S, Fujiwara M, Kondo H, Nishimiya Y, Yumoto I, Tsuda S (December 2003). "Antifreeze proteins from snow mold fungi". Canadian Journal of Botany. 81 (12): 1175–81. doi:10.1139/b03-116.
- Raymond JA, Janech MG (April 2009). "Ice-binding proteins from enoki and shiitake mushrooms". Cryobiology. 58 (2): 151–6. doi:10.1016/j.cryobiol.2008.11.009. PMID 19121299.
- Hanada Y, Nishimiya Y, Miura A, Tsuda S, Kondo H (August 2014). "Hyperactive antifreeze protein from an Antarctic sea ice bacterium Colwellia sp. has a compound ice-binding site without repetitive sequences". The FEBS Journal. 281 (16): 3576–90. doi:10.1111/febs.12878. PMID 24938370. S2CID 8388070.
- Do H, Kim SJ, Kim HJ, Lee JH (April 2014). "Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1". Acta Crystallographica. Section D, Biological Crystallography. 70 (Pt 4): 1061–73. doi:10.1107/S1399004714000996. PMID 24699650.
- Mangiagalli M, Bar-Dolev M, Tedesco P, Natalello A, Kaleda A, Brocca S, et al. (January 2017). "Cryo-protective effect of an ice-binding protein derived from Antarctic bacteria". The FEBS Journal. 284 (1): 163–177. doi:10.1111/febs.13965. hdl:11581/397803. PMID 27860412. S2CID 43854468.
- Barker PF, Thomas E (June 2004). "Origin, signature and palaeoclimatic influence of the Antarctic Circumpolar Current". Earth-Science Reviews. 66 (1): 143–162. Bibcode:2004ESRv...66..143B. doi:10.1016/j.earscirev.2003.10.003. ISSN 0012-8252.
- Eastman JT (January 2005). "The nature of the diversity of Antarctic fishes". Polar Biology. 28 (2): 93–107. doi:10.1007/s00300-004-0667-4. ISSN 1432-2056. S2CID 1653548.
- Chen L, DeVries AL, Cheng CH (April 1997). "Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod". Proceedings of the National Academy of Sciences of the United States of America. 94 (8): 3817–3822. Bibcode:1997PNAS...94.3817C. doi:10.1073/pnas.94.8.3817. PMC 20524. PMID 9108061.
- Wang, Sen; Amornwittawat, Natapol; Juwita, Vonny; Kao, Yu; Duman, John G.; Pascal, Tod A.; Goddard, William A.; Wen, Xin (2009-10-13). "Arginine, a Key Residue for the Enhancing Ability of an Antifreeze Protein of the Beetle Dendroides canadensis". Biochemistry. 48 (40): 9696–9703. doi:10.1021/bi901283p. ISSN 0006-2960. PMC 2760095. PMID 19746966.
- Wang, Lei; Duman, John G. (2006-01-31). "A thaumatin-like protein from larvae of the beetle Dendroides canadensis enhances the activity of antifreeze proteins". Biochemistry. 45 (4): 1278–1284. doi:10.1021/bi051680r. ISSN 0006-2960. PMID 16430224.
- Amornwittawat, Natapol; Wang, Sen; Duman, John G.; Wen, Xin (December 2008). "Polycarboxylates Enhance Beetle Antifreeze Protein Activity". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1784 (12): 1942–1948. doi:10.1016/j.bbapap.2008.06.003. ISSN 0006-3002. PMC 2632549. PMID 18620083.
- Li, N.; Andorfer, C. A.; Duman, J. G. (August 1998). "Enhancement of insect antifreeze protein activity by solutes of low molecular mass". The Journal of Experimental Biology. 201 (Pt 15): 2243–2251. doi:10.1242/jeb.201.15.2243. ISSN 0022-0949. PMID 9662495.
- Graham LA, Davies PL (June 2021). "Horizontal Gene Transfer in Vertebrates: A Fishy Tale". Trends in Genetics. 37 (6): 501–503. doi:10.1016/j.tig.2021.02.006. ISSN 0168-9525. PMID 33714557. S2CID 232232148.
- Chen L, DeVries AL, Cheng CH (April 1997). "Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod". Proceedings of the National Academy of Sciences of the United States of America. 94 (8): 3817–22. Bibcode:1997PNAS...94.3817C. doi:10.1073/pnas.94.8.3817. PMC 20524. PMID 9108061.
- Raymond JA, Lin Y, DeVries AL (July 1975). "Glycoprotein and protein antifreezes in two Alaskan fishes". The Journal of Experimental Zoology. 193 (1): 125–130. doi:10.1002/jez.1401930112. PMID 1141843.
- Hargens AR (April 1972). "Freezing resistance in polar fishes". Science. 176 (4031): 184–186. Bibcode:1972Sci...176..184H. doi:10.1126/science.176.4031.184. PMID 17843537. S2CID 45112534.
- Feeney RE, Yeh Y (1978-01-01). Anfinsen CB, Edsall JT, Richards FM (eds.). "Antifreeze proteins from fish bloods". Advances in Protein Chemistry. Academic Press. 32: 191–282. doi:10.1016/s0065-3233(08)60576-8. ISBN 9780120342327. PMID 362870.
- Graether, Steffen P.; Sykes, Brian D. (2004-07-14). "Cold survival in freeze-intolerant insects: The structure and function of β-helical antifreeze proteins". European Journal of Biochemistry. 271 (16): 3285–3296. doi:10.1111/j.1432-1033.2004.04256.x. PMID 15291806.
- Xiao, Nan; Suzuki, Keita; Nishimiya, Yoshiyuki; Kondo, Hidemasa; Miura, Ai; Tsuda, Sakae; Hoshino, Tamotsu (January 2010). "Comparison of functional properties of two fungal antifreeze proteins from Antarctomyces psychrotrophicus and Typhula ishikariensis: Antifreeze protein from ascomycetous fungus". FEBS Journal. 277 (2): 394–403. doi:10.1111/j.1742-4658.2009.07490.x. PMID 20030710. S2CID 3529668.
- Raymond JA, DeVries AL (June 1977). "Adsorption inhibition as a mechanism of freezing resistance in polar fishes". Proceedings of the National Academy of Sciences of the United States of America. 74 (6): 2589–93. Bibcode:1977PNAS...74.2589R. doi:10.1073/pnas.74.6.2589. PMC 432219. PMID 267952.
- Raymond JA, Wilson P, DeVries AL (February 1989). "Inhibition of growth of nonbasal planes in ice by fish antifreezes". Proceedings of the National Academy of Sciences of the United States of America. 86 (3): 881–5. Bibcode:1989PNAS...86..881R. doi:10.1073/pnas.86.3.881. PMC 286582. PMID 2915983.
- Yang DS, Hon WC, Bubanko S, Xue Y, Seetharaman J, Hew CL, Sicheri F (May 1998). "Identification of the ice-binding surface on a type III antifreeze protein with a "flatness function" algorithm". Biophysical Journal. 74 (5): 2142–51. Bibcode:1998BpJ....74.2142Y. doi:10.1016/S0006-3495(98)77923-8. PMC 1299557. PMID 9591641.
- Knight CA, Cheng CC, DeVries AL (February 1991). "Adsorption of alpha-helical antifreeze peptides on specific ice crystal surface planes". Biophysical Journal. 59 (2): 409–18. Bibcode:1991BpJ....59..409K. doi:10.1016/S0006-3495(91)82234-2. PMC 1281157. PMID 2009357.
- Haymet AD, Ward LG, Harding MM, Knight CA (July 1998). "Valine substituted winter flounder 'antifreeze': preservation of ice growth hysteresis". FEBS Letters. 430 (3): 301–6. doi:10.1016/S0014-5793(98)00652-8. PMID 9688560. S2CID 42371841.
- Chou KC (January 1992). "Energy-optimized structure of antifreeze protein and its binding mechanism". Journal of Molecular Biology. 223 (2): 509–17. doi:10.1016/0022-2836(92)90666-8. PMID 1738160.
- DeVries AL, Wohlschlag DE (March 1969). "Freezing resistance in some Antarctic fishes". Science. 163 (3871): 1073–5. Bibcode:1969Sci...163.1073D. doi:10.1126/science.163.3871.1073. PMID 5764871. S2CID 42048517.
- DeVries AL, Komatsu SK, Feeney RE (June 1970). "Chemical and physical properties of freezing point-depressing glycoproteins from Antarctic fishes". The Journal of Biological Chemistry. 245 (11): 2901–8. doi:10.1016/S0021-9258(18)63073-X. PMID 5488456.
- Urrutia ME, Duman JG, Knight CA (May 1992). "Plant thermal hysteresis proteins". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1121 (1–2): 199–206. doi:10.1016/0167-4838(92)90355-h. PMID 1599942.
- Duman JG, Olsen TM (1993). "Thermal hysteresis protein activity in bacteria, fungi and phylogenetically diverse plants". Cryobiology. 30 (3): 322–328. doi:10.1006/cryo.1993.1031.
- Clarke CJ, Buckley SL, Lindner N (2002). "Ice structuring proteins - a new name for antifreeze proteins". Cryo Letters. 23 (2): 89–92. PMID 12050776.
- Bressanini D. "Gelato OGM. Ma quando mai! Anche il formaggio allora..." Scienza in cucina. L'Espresso. Retrieved 6 July 2022.
- Merrett N (31 July 2007). "Unilever protein gets UK go ahead". DairyReporter.
- Thorington R (18 September 2014). "Can ice cream be tasty and healthy?". Impact Magazine. University of Nottingham.
- Dortch E (2006). "Fishy GM yeast used to make ice-cream". Network of Concerned Farmers. Archived from the original on 14 July 2011. Retrieved 9 October 2006.
- Moskin J (26 July 2006). "Creamy, Healthier Ice Cream? What's the Catch?". The New York Times.
- Regand A, Goff HD (January 2006). "Ice recrystallization inhibition in ice cream as affected by ice structuring proteins from winter wheat grass". Journal of Dairy Science. 89 (1): 49–57. doi:10.3168/jds.S0022-0302(06)72068-9. PMID 16357267.
- Celik Y, Graham LA, Mok YF, Bar M, Davies PL, Braslavsky I (2010). "Superheating of Ice in the Presence of Ice Binding Proteins". Biophysical Journal. 98 (3): 245a. Bibcode:2010BpJ....98..245C. doi:10.1016/j.bpj.2009.12.1331.*Lay summary in: "Antifreeze proteins can stop ice melt, new study finds". Physorg.com. March 1, 2010.
- Marc C (24 June 2021). "Des virus pour imiter les protéines antigel/".
Further reading
- Haymet AD, Ward LG, Harding MM (1999). "Winter Flounder 'anti-freeze' proteins: Synthesis and ice growth inhibition of analogues that probe the relative importance of hydrophobic and hydrogen bonding interactions". Journal of the American Chemical Society. 121 (5): 941–948. doi:10.1021/ja9801341. ISSN 0002-7863.
- Sicheri F, Yang DS (June 1995). "Ice-binding structure and mechanism of an antifreeze protein from winter flounder". Nature. 375 (6530): 427–31. Bibcode:1995Natur.375..427S. doi:10.1038/375427a0. hdl:11375/7005. PMID 7760940. S2CID 758990.
External links
- Cold, Hard Fact: Fish Antifreeze Produced in Pancreas
- Antifreeze Proteins: Molecule of the Month Archived 2015-11-04 at the Wayback Machine, by David Goodsell, RCSB Protein Data Bank
- Overview of all the structural information available in the PDB for UniProt: Q9GTP0 (Thermal hysteresis or Antifreeze protein) at the PDBe-KB.