Arginine decarboxylase
The enzyme Acid-Induced Arginine Decarboxylase (AdiA) (EC 4.1.1.19), also commonly referred to as arginine decarboxylase, catalyzes the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase.[1] It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH.[2] Arginine decarboxylase is part of an enzymatic system in Escherichia coli (E. coli),[3] Salmonella Typhimurium,[4] and methane-producing bacteria Methanococcus jannaschii[5] that makes these organisms acid resistant and allows them to survive under highly acidic medium.
| arginine decarboxylase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
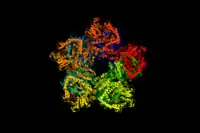 A 3d depiction of the arginine decarboxylase pentamer of homodimers | |||||||||
| Identifiers | |||||||||
| EC no. | 4.1.1.19 | ||||||||
| CAS no. | 9024-77-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Structure
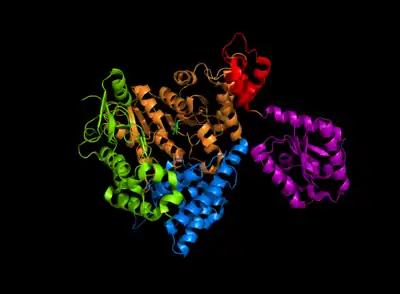
Arginine decarboxylase is a multimer of protein subunits. For instance, the form of this enzyme in E. coli is a ca. 800 kDa decamer of identical subunits, and is composed as a pentamer of dimers.[6] Each subunit can be divided into five domains: (1) the amino-terminal wing domain, (2) the linker domain, (3) the PLP-binding domain, (4) the aspartate aminotransferase- (AspAT-) like small domain, and (5) the carboxy-terminal domain.[3] The AspAT-like small domain, the PLP-binding domain, and the carboxy-terminal domain form an open bowl-like structure. The wing domain extends from the other three domains like a handle of the bowl, and the linker domain connects these two parts together. Altogether, the five domains associate with one another through hydrogen bonds and electrostatic interactions.[3]
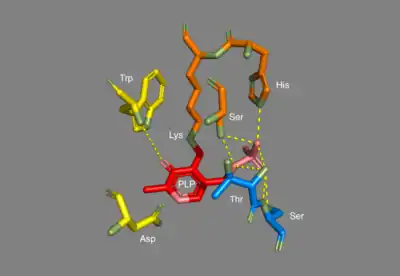
In E. coli arginine decarboxylase, each homodimer has two active sites that are buried about 30 Å from the dimer surface. The active site, found in the PLP-binding domain, consists of the PLP cofactor bound to a lysine residue in the form of a Schiff base. The phosphate group of PLP is held in place through hydrogen bonding with the alcoholic side chains of several serine and threonine residues, as well as through hydrogen bonding with the imidazole side chain of a histidine residue. The protonated nitrogen on the PLP aromatic ring is hydrogen bonded to a carboxylate on an aspartic side chain.[3]
Mechanism
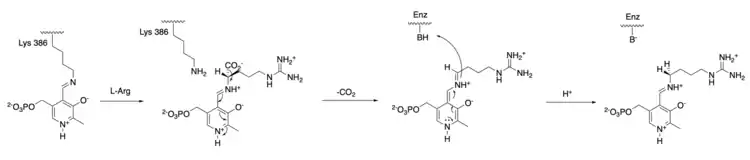
The mechanism of arginine decarboxylase is analogous to other deaminating and decarboxylating PLP enzymes in its use of a Schiff base intermediate.[7] Initially, Lys386 residue is displaced in a transamination reaction by the L-arginine substate, forming an arginine Schiff base with the PLP cofactor.[8] Decarboxylation of arginine carboxylate group then occurs, where it is hypothesized that the C-C bond broken is perpendicular to the PLP pyridine ring.[9] The pyridine nitrogen group acts as an electron-withdrawing group that facilitates the C-C bond breaking. Protonation of the amino acid leads to the formation of a new Schiff base that subsequently undergoes a transamination reaction by the lysine reside of arginine decarboxylase, regenerating the catalytically active PLP and releasing agmantine as a product. Though it has been hypothesized that a protonated histidine residue involves in the protonation step as a proton source,[10] the identity of the proton-donating residue in arginine decarboxylase has yet to be confirmed.
Function
Arginine decarboxylase is one of the main components of arginine-dependent acid resistance (AR3)[11] that allows E. coli to survive long enough in the highly acidic environment of the stomach to pass through the digestive tract and infect a human host. The enzyme consumes a cytoplasmic proton in the decarboxylation reaction, preventing the pH of the cell from becoming too acidic. The activity of the enzyme is dependent upon the surrounding pH. At more basic cellular pH levels, the enzyme exists in an inactive homodimer form, as electrostatic repulsion between negatively-charged acidic residues in the wing domains prevent homodimers from assembling into the catalytically active decamer. As the cellular environment becomes more acidic, these residues become neutrally charged through protonation. With less electrostatic repulsion between homodimers, the enzyme is allowed to assemble as the catalytically active decamer.[12] This particular assembling strategy used by E. coli arginine decarboxylase is also commonly used by other acidophilic organisms to cope with acidic growth conditions.[13] Overall, the acid resistance activity of arginine decarboxylase is two-fold. The surface protein residues of the homodimer consume protons, leading to the formation of active decamers which further increase proton consumption via the decarboxylation reaction. Arginine decarboxylase works in tandem with arginine decarboxylase antiporters (AdiC), another component of AR3, that exchange the extracellular arginine substrate for the intracellular by-product of decarboxylation.[14][15]
References
- Paiardini A, Contestabile R, Buckle AM, Cellini B (2014). "PLP-dependent enzymes". BioMed Research International. 2014: 856076. doi:10.1155/2014/856076. PMC 3914556. PMID 24527459.
- Boeker EA, Snell EE (January 1971). "[225] Arginine decarboxylase (Escherichia coli B)". [225] Arginine decarboxylase (Escherichia coli B). Methods in Enzymology. Vol. 17. pp. 657–662. doi:10.1016/0076-6879(71)17114-5. ISBN 9780121818777.
- Andréll J, Hicks MG, Palmer T, Carpenter EP, Iwata S, Maher MJ (May 2009). "Crystal structure of the acid-induced arginine decarboxylase from Escherichia coli: reversible decamer assembly controls enzyme activity". Biochemistry. 48 (18): 3915–27. doi:10.1021/bi900075d. PMID 19298070.
- Deka G, Bharath SR, Savithri HS, Murthy MR (September 2017). "Structural studies on the decameric S. Typhimurium arginine decarboxylase (ADC): Pyridoxal 5'-phosphate binding induces conformational changes". Biochemical and Biophysical Research Communications. 490 (4): 1362–1368. doi:10.1016/j.bbrc.2017.07.032. PMID 28694189.
- PDB: 1MT1; Tolbert WD, Graham DE, White RH, Ealick SE (March 2003). "Pyruvoyl-dependent arginine decarboxylase from Methanococcus jannaschii: crystal structures of the self-cleaved and S53A proenzyme forms". Structure. 11 (3): 285–94. doi:10.1016/S0969-2126(03)00026-1. PMID 12623016.
- Boeker EA, Snell EE (April 1968). "Arginine decarboxylase from Escherichia coli. II. Dissociation and reassociation of subunits". The Journal of Biological Chemistry. 243 (8): 1678–84. PMID 4870600.
- Eliot AC, Kirsch JF (2004). "Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations". Annual Review of Biochemistry. 73: 383–415. doi:10.1146/annurev.biochem.73.011303.074021. PMID 15189147.
- John RA (April 1995). "Pyridoxal phosphate-dependent enzymes". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1248 (2): 81–96. doi:10.1016/0167-4838(95)00025-p. PMID 7748903.
- Toney MD (January 2005). "Reaction specificity in pyridoxal phosphate enzymes". Archives of Biochemistry and Biophysics. 433 (1): 279–87. doi:10.1016/j.abb.2004.09.037. PMID 15581583.
- Akhtar M, Stevenson DE, Gani D (August 1990). "Fern L-methionine decarboxylase: kinetics and mechanism of decarboxylation and abortive transamination". Biochemistry. 29 (33): 7648–60. doi:10.1021/bi00485a014. PMID 2271524.
- Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW (September 1996). "Mechanisms of acid resistance in enterohemorrhagic Escherichia coli". Applied and Environmental Microbiology. 62 (9): 3094–100. doi:10.1128/AEM.62.9.3094-3100.1996. PMC 168100. PMID 8795195.
- Nowak S, Boeker EA (March 1981). "The inducible arginine decarboxylase of Escherichia coli B: activity of the dimer and the decamer". Archives of Biochemistry and Biophysics. 207 (1): 110–6. doi:10.1016/0003-9861(81)90015-1. PMID 7016035.
- Richard H, Foster JW (September 2004). "Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential". Journal of Bacteriology. 186 (18): 6032–41. doi:10.1128/jb.186.18.6032-6041.2004. PMC 515135. PMID 15342572.
- Gong S, Richard H, Foster JW (August 2003). "YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli". Journal of Bacteriology. 185 (15): 4402–9. doi:10.1128/jb.185.15.4402-4409.2003. PMC 165756. PMID 12867448.
- Iyer R, Williams C, Miller C (November 2003). "Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli". Journal of Bacteriology. 185 (22): 6556–61. doi:10.1128/jb.185.22.6556-6561.2003. PMC 262112. PMID 14594828.