Autolysin
Autolysins are endogenous lytic enzymes that break down the peptidoglycan components of biological cells which enables the separation of daughter cells following cell division.[1][2][3][4][5] They are involved in cell growth, cell wall metabolism, cell division and separation, as well as peptidoglycan turnover and have similar functions to lysozymes.[6]
Autolysin is formed from the precursor gene, Atl. Amidases (EC 3.5.1.28), gametolysin (EC 3.4.24.38), and glucosaminidase are considered as types of autolysins.[4][6]
Function and mechanisms
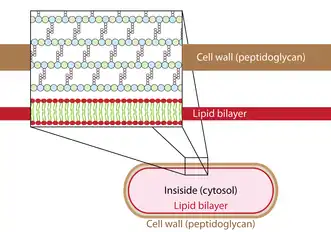
Autolysins exist in all bacteria containing peptidoglycan and are potentially considered as lethal enzymes when uncontrolled.[7] They target the glycosidic bonds as well as the cross-linked peptides of the peptidoglycan matrix.[8] The peptidoglycan matrix functions for cell wall stability to protect from turgor changes and carries out function for immunological defense.[9][10] These enzymes break down the peptidoglycan matrix in small sections to allow for peptidoglycan biosynthesis.[4] Autolysins breaks down old peptidoglycan which allows for the formation of newer peptidoglycan for cell growth and elongation. This is called cell wall turnover.[6] Autolysins do this by hydrolyzing the β-(1,4) glycosidic bond of the peptidoglycan cell wall and the linkage between N-acetylmuramoyl residues and L-amino acid residues of certain cell-wall glycopeptides.[4] This enzyme catalyses the following chemical reaction:
- Cleavage of the proline- and hydroxyproline-rich proteins of the Chlamydomonas cell wall; also cleaves azocasein, gelatin and Leu-Trp-Met-Arg-Phe-Ala
This glycoprotein is present in Chlamydomonas reinhardtii gametes.
Gram-positive bacteria regulate autolysins with teichoic acid molecules attached to the tetrapeptide of the peptidoglycan matrix.
The antibiotics complestatin and corbomycin prevent autolysin from remodeling the cell wall by binding to peptidoglycan, therefore stopping bacterial growth.[11] The amide linkages between stem peptide and lactyl moiety of muramoyl residue are cleaved by N-acetylmuramoyl-l-alanine amidases and partakes in cell separation and the dissociation of the cell septum.[4] There are 5 types of autolysins that contribute to cell separation of daughter cells, LytC, LytD, LytE, and LytF.[6]
In a study conducted with mice, those that were immunized with autolysin were able to survive longer than the infected mice. This study was able to support as evidence autolysin's contribution in virulence and potential for vaccine antigen.[12]
Lysis of mother cell
| N-acetylmuramoyl-L-alanine amidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.5.1.28 | ||||||||
| CAS no. | 9013-25-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
LytC and CwlC are two amidases from the LytC family that hydrolyze the peptidoglycan of the mother cell wall to allow for the release of the mature endospore. CwlC is directly found in the mother cell wall.[6]
Motility
Expression of lytC, lytD, and lytF genes together leads to flagellar motility and is controlled by the activity of the chemotaxis sigma factor, σD. The activity of this sigma factor peaks at the start of the stationary phase.[6]
Potential lethality
Autolysins are naturally produced by peptidoglycan containing bacteria, but excessive amounts will degrade the peptidoglycan matrix and cause the cell to burst due to osmotic pressure. Previous studies have found that the byproducts of autolysin during cell wall breakdown are highly immunogenic.[12] When observed in the bacteria, Bacillus subtilis, there were potentially lethal amounts of autolysin found in the cell walls.[6] In Streptococcus pneumoniae it was found that N-acetylmuramoyl-l-alanine amidase, a cell wall autolysin, could assist in pathogenesis due to its ability to break down the wall or lyse a portion of the invading pneumococci and release potentially lethal toxins into the cell. Researchers studied the function, structure, and cloning ability through Escherichia coli and also determined its nucleotide sequence.[12]
Families
LytC
LytC as well as LytD are considered as two major autolysins that contribute to vegetative cell wall growth and account for 95% of the autolytic activity in B. subtilis. LytC is found in the cell wall. LytB, a non-autolysin, was found to enhance LytC activity.[6] LytC and LytA interact and function together for lysis and cell death.[13]
| Gametolysin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.4.24.38 | ||||||||
| CAS no. | 97089-74-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
CwlC
CwlC is found in the mother cell wall and functions for the lysis of the mother cell wall.[6] CwlC does not have a signal sequence but participates in late sporulation and is present in the cell wall.[14][15] It was found in B. subtilis that CwlC is able to hydrolyze both vegetative cell walls and spore peptidoglycan.[14]
LytD glucosaminidase family
This family of autolysin consist of only LytD itself. LytD functions for vegetative growth. Autolytic activity is found within the C-terminal region with catalytic domain homologous to the glucosaminidase domain. LytD is found in the cell wall. LytD activity was studied in B. subtilis and glucosaminidase activity was found in mature glycan strands due to the presence of MurNAc at the nonreducing ends.[6]
See also
References
- Jaenicke L, Kuhne W, Spessert R, Wahle U, Waffenschmidt S (December 1987). "Cell-wall lytic enzymes (autolysins) of Chlamydomonas reinhardtii are (hydroxy)proline-specific proteases". European Journal of Biochemistry. 170 (1–2): 485–491. doi:10.1111/j.1432-1033.1987.tb13725.x. PMID 3319620.
- Buchanan MJ, Imam SH, Eskue WA, Snell WJ (January 1989). "Activation of the cell wall degrading protease, lysin, during sexual signalling in Chlamydomonas: the enzyme is stored as an inactive, higher relative molecular mass precursor in the periplasm". The Journal of Cell Biology. 108 (1): 199–207. doi:10.1083/jcb.108.1.199. PMC 2115355. PMID 2910877.
- Matsuda Y (1998). "Gametolysin". In Barrett AJ, Rawlings ND, Woessner JF (eds.). Handbook of Proteolytic Enzymes. London: Academic Press. pp. 1140–1143.
- Clarke AJ (September 2018). "The "hole" story of predatory outer-membrane vesicles". Canadian Journal of Microbiology. 64 (9): 589–599. doi:10.1139/cjm-2017-0466. PMID 30169125.
- Porayath C, Suresh MK, Biswas R, Nair BG, Mishra N, Pal S (April 2018). "Autolysin mediated adherence of Staphylococcus aureus with Fibronectin, Gelatin and Heparin". International Journal of Biological Macromolecules. 110: 179–184. doi:10.1016/j.ijbiomac.2018.01.047. PMC 5864509. PMID 29398086.
- Smith TJ, Blackman SA, Foster SJ (February 2000). "Autolysins of Bacillus subtilis: multiple enzymes with multiple functions". Microbiology. 146 ( Pt 2) (2): 249–262. doi:10.1099/00221287-146-2-249. PMID 10708363.
- Haghighat S, Siadat SD, Sorkhabadi SM, Sepahi AA, Mahdavi M (February 2017). "Cloning, expression and purification of autolysin from methicillin-resistant Staphylococcus aureus: potency and challenge study in Balb/c mice". Molecular Immunology. 82: 10–18. doi:10.1016/j.molimm.2016.12.013. PMID 28006655. S2CID 36593600.
- Atilano ML, Pereira PM, Vaz F, Catalão MJ, Reed P, Grilo IR, et al. (April 2014). "Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system". eLife. 3: e02277. doi:10.7554/eLife.02277. PMC 3971415. PMID 24692449.
- Zhang Y, Zhong X, Lu P, Zhu Y, Dong W, Roy S, et al. (July 2019). "A novel autolysin AtlASS mediates bacterial cell separation during cell division and contributes to full virulence in Streptococcus suis". Veterinary Microbiology. 234: 92–100. doi:10.1016/j.vetmic.2019.05.020. PMID 31213278. S2CID 182382454.
- Pazos M, Peters K (2019). "Peptidoglycan". In Kuhn A (ed.). Bacterial Cell Walls and Membranes. Subcellular Biochemistry. Vol. 92. Springer International Publishing. pp. 127–168. doi:10.1007/978-3-030-18768-2_5. ISBN 978-3-030-18767-5. PMID 31214986. S2CID 239142525.
- Culp EJ, Waglechner N, Wang W, Fiebig-Comyn AA, Hsu YP, Koteva K, et al. (February 2020). "Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling". Nature. 578 (7796): 582–587. Bibcode:2020Natur.578..582C. doi:10.1038/s41586-020-1990-9. PMID 32051588. S2CID 211089119.
- Berry AM, Lock RA, Hansman D, Paton JC (August 1989). "Contribution of autolysin to virulence of Streptococcus pneumoniae". Infection and Immunity. 57 (8): 2324–2330. doi:10.1128/iai.57.8.2324-2330.1989. PMC 313450. PMID 2568343.
- García P, Paz González M, García E, García JL, López R (July 1999). "The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains". Molecular Microbiology. 33 (1): 128–138. doi:10.1046/j.1365-2958.1999.01455.x. PMID 10411730.
- Kuroda A, Asami Y, Sekiguchi J (October 1993). "Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis". Journal of Bacteriology. 175 (19): 6260–6268. doi:10.1128/jb.175.19.6260-6268.1993. PMC 206722. PMID 8407798.
- Smith TJ, Foster SJ (July 1995). "Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168". Journal of Bacteriology. 177 (13): 3855–3862. doi:10.1128/jb.177.13.3855-3862.1995. PMC 177106. PMID 7601853.
External links
- Gametolysin at the U.S. National Library of Medicine Medical Subject Headings (MeSH)