Chlamydomonas reinhardtii
Chlamydomonas reinhardtii is a single-cell green alga about 10 micrometres in diameter that swims with two flagella. It has a cell wall made of hydroxyproline-rich glycoproteins, a large cup-shaped chloroplast, a large pyrenoid, and an eyespot that senses light.
| Chlamydomonas reinhardtii | |
|---|---|
 | |
| Scientific classification | |
| (unranked): | Viridiplantae |
| Division: | Chlorophyta |
| Class: | Chlorophyceae |
| Order: | Chlamydomonadales |
| Family: | Chlamydomonadaceae |
| Genus: | Chlamydomonas |
| Species: | C. reinhardtii |
| Binomial name | |
| Chlamydomonas reinhardtii | |
Chlamydomonas species are widely distributed worldwide in soil and fresh water. Chlamydomonas reinhardtii is an especially well studied biological model organism, partly due to its ease of culturing and the ability to manipulate its genetics. When illuminated, C. reinhardtii can grow photoautotrophically, but it can also grow in the dark if supplied with organic carbon. Commercially, C. reinhardtii is of interest for producing biopharmaceuticals and biofuel, as well being a valuable research tool in making hydrogen.
History
The C. reinhardtii wild-type laboratory strain c137 (mt+) originates from an isolate collected near Amherst, Massachusetts, in 1945 by Gilbert M. Smith.[1][2]
The species' name has been spelled several different ways because of different transliterations of the name from Russian: reinhardi, reinhardii, and reinhardtii all refer to the same species, C. reinhardtii Dangeard.[3]
Model organism

Chlamydomonas is used as a model organism for research on fundamental questions in cell and molecular biology such as:
- How do cells move?
- How do cells respond to light?
- How do cells recognize one another?
- How do cells generate regular, repeatable flagellar waveforms?
- How do cells regulate their proteome to control flagellar length?
- How do cells respond to changes in mineral nutrition? (nitrogen, sulfur, etc.)
There are many known mutants of C. reinhardtii. These mutants are useful tools for studying a variety of biological processes, including flagellar motility, photosynthesis, and protein synthesis. Since Chlamydomonas species are normally haploid, the effects of mutations are seen immediately without further crosses.
In 2007, the complete nuclear genome sequence of C. reinhardtii was published.[4]
Channelrhodopsin-1 and Channelrhodopsin-2, proteins that function as light-gated cation channels, were originally isolated from C. reinhardtii.[5][6] These proteins and others like them are increasingly widely used in the field of optogenetics.[7]
Mitochondrial significance
The genome of C. reinhardtii is significant for mitochondrial study as it is one species where the genes for 6 of the 13 proteins encoded for the mitochondria are found in the nucleus of the cell, leaving 7 in the mitochondria. In all other species these genes are present only in the mitochondria and are unable to be allotopically expressed. This is significant for the testing and development of therapies for genetic mitochondrial diseases.
Reproduction
Vegetative cells of reinhardtii species are haploid with 17 small chromosomes. Under nitrogen starvation, vegetative cells differentiate into haploid gametes.[8] There are two mating types, identical in appearance, thus isogamous, and known as mt(+) and mt(-), which can fuse to form a diploid zygote. The zygote is not flagellated, and it serves as a dormant form of the species in the soil. In the light, the zygote undergoes meiosis and releases four flagellated haploid cells that resume the vegetative lifecycle.
Under ideal growth conditions, cells may sometimes undergo two or three rounds of mitosis before the daughter cells are released from the old cell wall into the medium. Thus, a single growth step may result in 4 or 8 daughter cells per mother cell.
The cell cycle of this unicellular green algae can be synchronized by alternating periods of light and dark. The growth phase is dependent on light, whereas, after a point designated as the transition or commitment point, processes are light-independent.[9]
Eye spot
C. reinhardtii has an eyespot similar to that of dinoflagellates.[10] The eyespot is located near the cell equator. It is composed of a carotenoid-rich granule layer in the chloroplast which act like a light reflector.[11] The main function of the eyespot is the phototaxis, which consist of the movement (with the flagella) related to a light stimulus.[12] The phototaxis is crucial for the alga and allows for localization of the environment with optimal light conditions for photosynthesis.[13] Phototaxis can be positive or negative depending on the light intensity.[10] The phototactic pathway consists of four steps leading to a change in the beating balance between the two flagella (the cis-flagellum which is the one closest to the eyespot, and the trans-flagellum which is the one farthest from the eyespot).[12]
Genetics
The attractiveness of the algae as a model organism has recently increased with the release of several genomic resources to the public domain. The Chlre3 draft of the Chlamydomonas nuclear genome sequence prepared by Joint Genome Institute of the U.S. Dept of Energy comprises 1557 scaffolds totaling 120 Mb. Roughly half of the genome is contained in 24 scaffolds all at least 1.6 Mb in length. The current assembly of the nuclear genome is available online.[14]
The ~15.8 Kb mitochondrial genome (database accession: NC_001638) is available online at the NCBI database.[15] The complete ~203.8 Kb chloroplast genome (database accession: NC_005353) is available online.[16][17]
In addition to genomic sequence data, there is a large supply of expression sequence data available as cDNA libraries and expressed sequence tags (ESTs). Seven cDNA libraries are available online.[18] A BAC library can be purchased from the Clemson University Genomics Institute.[19] There are also two databases of >50 000[20] and >160 000[21] ESTs available online.
A genome-wide collection of mutants with mapped insertion sites covering most nuclear genes[22][23] is available: https://www.chlamylibrary.org/.
The genome of C. reinhardtii has been shown to contain N6-Methyldeoxyadenosine (6mA), a mark common in prokaryotes but much rarer in eukaryotes.[24] Some research has indicated that 6mA in Chlamydomonas may be involved in nucleosome positioning, as it is present in the linker regions between nucleosomes as well as near the transcription start sites of actively transcribed genes.[25]
C. reinhardtii appears to be capable of several DNA repair processes.[26] These include recombinational repair, strand break repair and excision repair.
Experimental evolution
Chlamydomonas has been used to study different aspects of evolutionary biology and ecology. It is an organism of choice for many selection experiments because (1) it has a short generation time, (2) it is both an autotroph and a facultative heterotroph, (3) it can reproduce both sexually and asexually, and (4) there is a wealth of genetic information already available.
Some examples (nonexhaustive) of evolutionary work done with Chlamydomonas include the evolution of sexual reproduction,[27] the fitness effect of mutations,[28] and the effect of adaptation to different levels of CO2.[29]
According to one frequently cited theoretical hypothesis,[30] sexual reproduction (in contrast to asexual reproduction) is adaptively maintained in benign environments because it reduces mutational load by combining deleterious mutations from different lines of descent and increases mean fitness. However, in a long-term experimental study of C. reinhardtii, evidence was obtained that contradicted this hypothesis. In sexual populations, mutation clearance was not found to occur and fitness was not found to increase.[31]
Motion
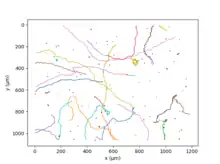
C. reinhardtii swims thanks to its two flagella,[32] in a movement analogous to human breaststroke. Repeating this elementary movement 50 times per second the algae have a mean velocity of 70 µm/s;[33] the genetic diversity of the different strains results in a huge range of values for this quantity. After few seconds of run, an asynchronous beating of the two flagella leads to a random change of direction, a movement called "run and tumble".[32] At a larger time and space scale, the random movement of the alga can be described as an active diffusion phenomenon.[34]
DNA transformation techniques
Gene transformation occurs mainly by homologous recombination in the chloroplast and heterologous recombination in the nucleus. The C. reinhardtii chloroplast genome can be transformed using microprojectile particle bombardment or glass bead agitation, however this last method is far less efficient. The nuclear genome has been transformed with both glass bead agitation and electroporation. The biolistic procedure appears to be the most efficient way of introducing DNA into the chloroplast genome. This is probably because the chloroplast occupies over half of the volume of the cell providing the microprojectile with a large target. Electroporation has been shown to be the most efficient way of introducing DNA into the nuclear genome with maximum transformation frequencies two orders of magnitude higher than obtained using glass bead method.
Production of biopharmaceuticals
Genetically engineered C. reinhardtii has been used to produce a mammalian serum amyloid protein (needs citation), a human antibody protein (needs citation), human Vascular endothelial growth factor, a potential therapeutic Human Papillomavirus 16 vaccine,[35] a potential malaria vaccine (an edible algae vaccine),[36] and a complex designer drug that could be used to treat cancer.[37]
Alternative protein source
C. reinhardtii is in production as a new algae-based nutritional source. Compared to Chlorella and Spirulina, C. reinhardtii was found to have more Alpha-linolenic acid, and a lower quantity of heavy metals while also containing all the essential amino acids and similar protein content.[38] Triton Algae Innovations is developing a commercial alternative protein product made from C reinhardtii.
Clean source of hydrogen production
In 1939, the German researcher Hans Gaffron (1902–1979), who was at that time attached to the University of Chicago, discovered the hydrogen metabolism of unicellular green algae. C reinhardtii and some other green algae can, under specified circumstances, stop producing oxygen and convert instead to the production of hydrogen. This reaction by hydrogenase, an enzyme active only in the absence of oxygen, is short-lived. Over the next thirty years, Gaffron and his team worked out the basic mechanics of this photosynthetic hydrogen production by algae.[39]
To increase the production of hydrogen, several tracks are being followed by the researchers.
- The first track is decoupling hydrogenase from photosynthesis. This way, oxygen accumulation can no longer inhibit the production of hydrogen. And, if one goes one step further by changing the structure of the enzyme hydrogenase, it becomes possible to render hydrogenase insensitive to oxygen. This makes a continuous production of hydrogen possible. In this case, the flux of electrons needed for this production no longer comes from the production of sugars but is drawn from the breakdown of its own stock of starch.[40]
- A second track is to interrupt temporarily, through genetic manipulation of hydrogenase, the photosynthesis process. This inhibits oxygen's reaching a level where it is able to stop the production of hydrogen.[41]
- The third track, mainly investigated by researchers in the 1950s, is chemical or mechanical methods of removal of O2 produced by the photosynthetic activity of the algal cells. These have included the addition of O2 scavengers, the use of added reductants, and purging the cultures with inert gases.[42] However, these methods are not inherently scalable, and may not be applicable to applied systems. New research has appeared on the subject of removing oxygen from algae cultures, and may eliminate scaling problems.
- The fourth track has been investigated, namely using copper salts to decouple hydrogenase action from oxygen production.[43]
- The fifth track has been suggested to reroute the photosynthetic electron flow from CO2 fixation in Calvin cycle to hydrogenase by applying short light pulses to anaerobic algae[44] or by depleting the culture of CO2.[45]
References
- "CC-125 wild type mt+ 137c". Chlamydomonas Center core collection list. Archived from the original on 2009-07-27. Retrieved 2009-03-09.
- The Chlamydomonas Sourcebook, ISBN 978-0-12-370873-1)
- http://megasun.bch.umontreal.ca/protists/chlamy/taxonomy.html Chlamydomonas Taxonomy.
- Merchant; Prochnik, SE; Vallon, O; Harris, EH; Karpowicz, SJ; Witman, GB; Terry, A; Salamov, A; et al. (2007). "The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions". Science. 318 (5848): 245–250. Bibcode:2007Sci...318..245M. doi:10.1126/science.1143609. PMC 2875087. PMID 17932292.
- Nagel G, Ollig D, Fuhrmann M, et al. (June 28, 2002). "Channelrhodopsin-1: a light-gated proton channel in green algae". Science. 296 (5577): 2395–8. Bibcode:2002Sci...296.2395N. doi:10.1126/science.1072068. PMID 12089443. S2CID 206506942.
- Lagali PS, Balya D, Awatramani GB, Münch TA, Kim DS, Busskamp V, Cepko CL, Roska B (June 2008). "Light-activated channels targeted to ON bipolar cells restore visual function in retinal degeneration". Nature Neuroscience. 11 (6): 667–75. doi:10.1038/nn.2117. PMID 18432197. S2CID 6798764.
- Boyden ES, et al. (May 3, 2011). "A history of optogenetics: the development of tools for controlling brain circuits with light". F1000 Biology Reports. 3 (11): 11. doi:10.3410/B3-11. PMC 3155186. PMID 21876722.
- SAGER R, GRANICK S (July 1954). "Nutritional control of sexuality in Chlamydomonas reinhardi". J. Gen. Physiol. 37 (6): 729–42. doi:10.1085/jgp.37.6.729. PMC 2147466. PMID 13174779.
- Oldenhof H.; Zachleder V.; den Ende H. (2006). "Blue- and red-light regulation of the cell cycle in Chlamydomonas reinhardtii (Chlorophyta)". Eur. J. Phycol. 41 (3): 313–320. doi:10.1080/09670260600699920.
- Ueki, Noriko; Ide, Takahiro; Mochiji, Shota; Kobayashi, Yuki; Tokutsu, Ryutaro; Ohnishi, Norikazu; Yamaguchi, Katsushi; Shigenobu, Shuji; Tanaka, Kan; Minagawa, Jun; Hisabori, Toru; Hirono, Masafumi; Wakabayashi, Ken-Ichi (2016). "Eyespot-dependent determination of the phototactic sign in Chlamydomonas reinhardtii". Proceedings of the National Academy of Sciences. 113 (19): 5299–5304. Bibcode:2016PNAS..113.5299U. doi:10.1073/pnas.1525538113. PMC 4868408. PMID 27122315.
- Foster, K.W. and Smyth, R.D. (1980) "Light Antennas in phototactic algae". Microbiological reviews, 44(4): 572–630.
- Hegemann P, Berthold P (2009) ""Sensory photoreceptors and light control of flagellar activity". In: Stern D, Witman G (Eds) The Chlamydomonas Sourcebook, second edition, volume 3, pages 395–430, Academic, Oxford. ISBN 9780123708731.
- Demmig-Adams, B.; Adams, W. W. (1992). "Photoprotection and Other Responses of Plants to High Light Stress". Annual Review of Plant Physiology and Plant Molecular Biology. 43: 599–626. doi:10.1146/annurev.pp.43.060192.003123.
- "Home - Chlamydomonas reinhardtii v3.0".
- "Chlamydomonas reinhardtii mitochondrion, complete genome". February 2010.
{{cite journal}}: Cite journal requires|journal=(help) - "Chlamydomonas reinhardtii chloroplast, complete genome". 2004-01-23.
{{cite journal}}: Cite journal requires|journal=(help) - "Chlamydomonas Chloroplast Genome Portal".
- "Chlamydomonas Center - Libraries". Archived from the original on 2004-10-19. Retrieved 2006-09-28.
- "CUGI". Archived from the original on 2014-12-26. Retrieved 2006-04-03.
- "[KDRI]Chlamydomonas reinhardtii EST index".
- "Search". Archived from the original on 2005-02-04. Retrieved 2006-09-28.
- Li, Xiaobo; Zhang, Ru; Patena, Weronika; Gang, Spencer S.; Blum, Sean R.; Ivanova, Nina; Yue, Rebecca; Robertson, Jacob M.; Lefebvre, Paul A.; Fitz-Gibbon, Sorel T.; Grossman, Arthur R.; Jonikas, Martin C. (2016-02-01). "An Indexed, Mapped Mutant Library Enables Reverse Genetics Studies of Biological Processes in Chlamydomonas reinhardtii". The Plant Cell. 28 (2): 367–387. doi:10.1105/tpc.15.00465. ISSN 1040-4651. PMC 4790863. PMID 26764374.
- Li, Xiaobo; Patena, Weronika; Fauser, Friedrich; Jinkerson, Robert E.; Saroussi, Shai; Meyer, Moritz T.; Ivanova, Nina; Robertson, Jacob M.; Yue, Rebecca; Zhang, Ru; Vilarrasa-Blasi, Josep; Wittkopp, Tyler M.; Ramundo, Silvia; Blum, Sean R.; Goh, Audrey; Laudon, Matthew; Srikumar, Tharan; Lefebvre, Paul A.; Grossman, Arthur R.; Jonikas, Martin C. (April 2019). "A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis". Nature Genetics. 51 (4): 627–635. doi:10.1038/s41588-019-0370-6. ISSN 1546-1718. PMC 6636631. PMID 30886426.
- Hattman, S; Kenny, C; Berger, L; Pratt, K (September 1978). "Comparative study of DNA methylation in three unicellular eucaryotes". Journal of Bacteriology. 135 (3): 1156–7. doi:10.1128/JB.135.3.1156-1157.1978. PMC 222496. PMID 99431.
- Fu, Ye; Luo, Guan-Zheng; Chen, Kai; Deng, Xin; Yu, Miao; Han, Dali; Hao, Ziyang; Liu, Jianzhao; Lu, Xingyu; Doré, Louis C.; Weng, Xiaocheng; Ji, Quanjiang; Mets, Laurens; He, Chuan (May 2015). "N6-Methyldeoxyadenosine Marks Active Transcription Start Sites in Chlamydomonas". Cell. 161 (4): 879–892. doi:10.1016/j.cell.2015.04.010. PMC 4427561. PMID 25936837.
- Vlcek D, Sevcovicová A, Sviezená B, Gálová E, Miadoková E. Chlamydomonas reinhardtii: a convenient model system for the study of DNA repair in photoautotrophic eukaryotes. Curr Genet. 2008 Jan;53(1):1-22. doi: 10.1007/s00294-007-0163-9. Epub 2007 Nov 9. PMID 17992532
- Colegrave N (2002). "Sex releases the speed limit on evolution". Nature. 420 (6916): 664–666. Bibcode:2002Natur.420..664C. doi:10.1038/nature01191. hdl:1842/692. PMID 12478292. S2CID 4382757.
- De Visser et al. 1996 The effect of sex and deleterious mutations on fitness in Chlamydomonas. Proc. R. Soc. Lond. B 263-193-200.
- Collins , Bell (2004). "Phenotypic consequences of 1,000 generations of selection at elevated CO2 in a green alga". Nature. 431 (7008): 566–569. Bibcode:2004Natur.431..566C. doi:10.1038/nature02945. PMID 15457260. S2CID 4354542.
- Kondrashov AS (October 1984). "Deleterious mutations as an evolutionary factor. 1. The advantage of recombination". Genet. Res. 44 (2): 199–217. doi:10.1017/s0016672300026392. PMID 6510714.
- Renaut S, Replansky T, Heppleston A, Bell G (November 2006). "The ecology and genetics of fitness in Chlamydomonas. XIII. Fitness of long-term sexual and asexual populations in benign environments". Evolution. 60 (11): 2272–9. doi:10.1554/06-084.1. PMID 17236420. S2CID 18977144.
- Polin, Marco; Tuval, Idan; Drescher, Knut; Gollub, J. P.; Goldstein, Raymond E. (2009-07-24). "Chlamydomonas Swims with Two "Gears" in a Eukaryotic Version of Run-and-Tumble Locomotion". Science. 325 (5939): 487–490. Bibcode:2009Sci...325..487P. doi:10.1126/science.1172667. ISSN 0036-8075. PMID 19628868. S2CID 10530835.
- Garcia, Michaël (2013-07-09). Hydrodynamique de micro-nageurs (phdthesis thesis) (in French). Université de Grenoble.
- Goldstein, Raymond E (2018-07-23). "Are theoretical results 'Results'?". eLife. 7: e40018. doi:10.7554/eLife.40018. ISSN 2050-084X. PMC 6056240. PMID 30033910.
- Demurtas OC; Massa S; Ferrante P; Venuti A; Franconi R; et al. (2013). "A Chlamydomonas-Derived Human Papillomavirus 16 E7 Vaccine Induces Specific Tumor Protection". PLOS ONE. 8 (4): e61473. Bibcode:2013PLoSO...861473D. doi:10.1371/journal.pone.0061473. PMC 3634004. PMID 23626690.
- (16 May 2012) Biologists produce potential malarial vaccine from algae PhysOrg, Retrieved 15 April 2013
- (10 December 2012) Engineering algae to make complex anti-cancer 'designer' drug PhysOrg, Retrieved 15 April 2013
- Darwish, Randa; Gedi, Mohamed; Akepach, Patchaniya; Assaye, Hirut; Zaky, Abderlahman; Gray, David (26 September 2020). "Chlamydomonas reinhardtii Is a Potential Food Supplement with the Capacity to Outperform Chlorella and Spirulina". Applied Sciences. 10 (19): 6736. doi:10.3390/app10196736. Retrieved 26 August 2021.
- Anastasios Melis; Thomas Happe (2004). "Trails of green alga hydrogen research — from Hans Gaffron to new frontiers" (PDF). Photosynthesis Research. 80 (1–3): 401–409. doi:10.1023/B:PRES.0000030421.31730.cb. PMID 16328836. S2CID 7188276.
- Laurent Cournac; Florence Musa; Laetitia Bernarda; Geneviève Guedeneya; Paulette Vignaisb; Gilles Peltie (2002). "Limiting steps of hydrogen production in Chlamydomonas reinhardtii and Synechocystis PCC 6803 as analysed by light-induced gas exchange transients". International Journal of Hydrogen Energy. 27 (11/12): 1229–1237. doi:10.1016/S0360-3199(02)00105-2.
- Anastasios Melis. "Hydrogen and hydrocarbon biofuels production via microalgal photosynthesis". Archived from the original on 2008-04-03. Retrieved 2008-04-07.
- Kosourov, S.; Tsyganov, A.; Seibert, M.; Ghirardi, M. (June 2002). "Sustained Hydrogen Photoproduction by Chlamydomonas reinhardtii:Effects of Culture Parameters". Biotechnol. Bioeng. 78 (7): 731–40. doi:10.1002/bit.10254. PMID 12001165.
- Fernandez VM, Rua ML, Reyes P, Cammack R, Hatchikian EC (November 1989). "Inhibition of Desulfovibrio gigas hydrogenase with copper salts and other metal ions". Eur. J. Biochem. 185 (2): 449–54. doi:10.1111/j.1432-1033.1989.tb15135.x. PMID 2555191.
- Kosourov, S.; Jokel, M.; Aro, E.-M.; Allahverdiyeva, Y. (March 2018). "A new approach for sustained and efficient H2 photoproduction by Chlamydomonas reinhardtii". Energy & Environmental Science. 11 (6): 1431–1436. doi:10.1039/C8EE00054A.
- Nagy, V.; Podmaniczki, A.; Vidal-Meireles, A.; Tengölics, R.; Kovács, L.; Rákhely, G.; Scoma, A.; Tóth SZ. (March 2018). "Water-splitting-based, sustainable and efficient H2 production in green algae as achieved by substrate limitation of the Calvin–Benson–Bassham cycle". Biotechnology for Biofuels. 11: 69. doi:10.1186/s13068-018-1069-0. PMC 5858145. PMID 29560024.
Further reading
Aoyama, H., Kuroiwa, T. and Nakamura, S. 2009. The dynamic behaviour of mitochondria in living zygotes during maturation and meiosis in Chlamydomonas reinhardtii. Eur. J. Phycol. 44: 497 - 507. doi:10.1080/09670260903272599
Jamers, A., Lenjou, M., Deraedt, P., van Bockstaele, D., Blust, R. and de Coen, W. 2009. Flow cytometric analysis of the cadmium-exposed green algae Chlamydomonas reinhadtii (Chlorophyceae). Eur. J. Phycol. 44: 541 - 550. doi:10.1080/09670260903118214
External links
- The Chlamydomonas Resource Center - "A central repository to receive, catalog, preserve, and distribute high-quality and reliable wild type and mutant cultures of the green alga Chlamydomonas reinhardtii, as well as useful molecular reagents and kits for education and research."
- Plant Comparative Genomics portal - Chlamydomonas reinhardtii resources from the Department of Energy Joint Genome Institute
- Guiry, M.D.; Guiry, G.M. "Chlamydomonas reinhardtii". AlgaeBase. World-wide electronic publication, National University of Ireland, Galway.
- Chlamydomonas reinhardtii cell, life cycle, strains, mating types - archived database.