Chitinase domain-containing protein 1
Chitinase domain-containing protein 1 (CHID1) is a highly conserved protein of unknown function located on the short (p) arm of chromosome 11 near the telomere.[5] The protein has 27 introns, which allows for many isoforms of this gene. It has several aliases, the most common of which is Stabilin-1 interacting chitinase-like protein (SI-CLP). As indicated by the alias, CHID1 is known to interact with the protein STAB1.[6] CHID1 is expressed ubiquitously at levels nearly 6 times the average gene,[7] and is conserved very far back to organisms such as Caenorhabditis elegans and possibly some prokaryotes. This protein is known to have carbohydrate binding sites, which could be involved in carbohydrate catabolysis.
| CHID1 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | CHID1, SI-CLP, SICLP, GL008, chitinase domain containing 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 615692 MGI: 1915288 HomoloGene: 12229 GeneCards: CHID1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Gene
CHID1 is located on chromosome 11 at the location p15.5.[8] It is just downstream of TSPAN4 and upstream of AP2A2.[9] CHID1 is ubiquitously expressed at a high levels. Through microarray analysis, it has been shown that CHID1 is generally expressed at 5.7 times the average gene.[7] CHID1 has many known variants, which is attributed to its 37 exons. There are no inherent repeats or hairpin structures to be found within the coding region of CHID1. This gene is a member of the GH18 superfamily, which dictates some of its protein structure. This gene also has several aliases, the most common of which is Stabilin-1 interacting chitinase-like protein, or SI-CLP, which indicates its known interaction with STAB1.[10]
Variations
Due to its large size and many exons, CHID1 has many transcript variants that have been identified through mRNA sequencing. CHID1 has 27 exons and 22 known splice forms.[7] These forms indicate that there may be multiple promotor regions and transcription start sites used within the genome. The most commonly found transcripts translate to about 400 amino acids each.
Homology
CHID1 shows a very high level of conservation. It has been identified in many model systems including Drosophila melanogaster, Caenorhabditis elegans, and Oryza sativa.[11] When conservation is observed over a large region, it becomes clear which protein domains are most important to CHID1. By far, the two best conserved factors are exon junctions you can add a link to exon junction, as well as several known carbohydrate binding sites. Another important region appears to be the last 15 amino acids, which retain a high level of conservation even through insect sequences.

| Species | Common name | Divergence in millions of years | Accession number | Similarity to human sequence |
|---|---|---|---|---|
| Gorilla gorilla gorilla | Gorilla | 8.8 | XP_004050445.1 | 100% |
| Macaca mulatta | Rhesus macaque | 29.0 | AFI38071.1 | 94% |
| Equus caballus | Horse | 94.2 | XP_003362692.1 | 91% |
| Felis catus | House cat | 94.2 | XP_003993852.1 | 90% |
| Taeniopygia guttata | Finch | 296 | XP_002199280.2 | 78% |
| Ictalurus punctatus | Catfish | 400.1 | NP_001187344.1 | 70% |
| Apis florea | Honey bee | 782.7 | XP_003691984.1 | 61% |
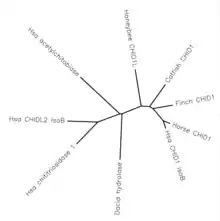
Paralogs
There are also several proposed paralogs to CHID1. These genes are di-N-acetylchitobiase, Chitinase-3-like protein 2, and chitotriosidase 1. All of these genes have roughly 40% similarity, and match human transcripts of CHID1 over 40-50% of the gene.[12] Based on the level of conservation, these paralogs all likely split at a time near to the split from bacteria.
| Name | Accession number | Length (amino acids) | Similarity to CHID1 in humans |
|---|---|---|---|
| di-N-acetylchitobiase | NP_004379.1 | 385 | 38% |
| Chitinase-3-like protein 2 | NP_001020368.1 | 380 | 39% |
| Chitotriosidase-1 | NP_001243054.2 | 447 | 40% |
Protein
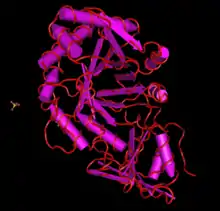
The protein translation of CHID1 is typically about 400 amino acids long (though this varies within the many known forms), and has few post-translational modifications.[13] In most forms and orthologs, it is predicted that CHID1 has a signal peptide that varies in length by sequence.[14] Structure predictions and x-ray crystallography structures of CHID1 indicate that its secondary structure is heavy in alpha helices, though it definitely has some beta strands present, as indicated by the SI-CLP x-ray crystallography structure. CHID1 is also a member of the GH18 superfamily of proteins, which has a unique conserved structure.[15] Members of this family contain an 8 stranded beta/alpha barrel. This protein is also shown to interact with other copies of itself in a homodimer, which also uses sulfate ion interacting molecules for an unknown purpose.
Expression

The expression of CHID1 is consistently shown to be ubiquitous and higher than average in all human tissues.[16][17][18] When analyzing the promoter region of CHID1 in humans, this expression is explained by the large number of transcription factor binding sites which are active in a wide range of tissues or are even ubiquitous.[19] In Drosophila melanogaster, tissue expression varies wildly between larval and adult tissue.[20] While expression is extremely widespread in adults, it is not at such a high level as in humans. In larvae, CHID1 is only shown to be expressed in specific tissues, and is up-regulated in very different tissues than in adults. In larval tissues, expression is highest in the salivary gland, but in adults the highest expression by far is in the male accessory gland.[20] Though expression is generally high in some species, it is not necessarily abnormal. In situ hybridization of gene transcripts in mouse brains shows that CHID1 is expressed relatively normally.[21] The pattern of high and low expression with the brain is very similar to another widespread gene: beta actin.[22] This is one indication that CHID1 may show normal expression patterning in mammals, despite its general upregulation.
Function
Although CHID1 has no known function for certain, there are several proposed activities of the protein based on current knowledge. This protein may participate in metabolic processes such as chitin catabolism or carbohydrate metabolism. It may locate in various cellular compartments such as the cytoplasm, or in lysosomes all depending on certain post-translational modifications.[23] Another study proposed that CHID1 may have roles in pathogen sensing.[15] The function of CHID1 may also depend on its putative binding partner, STAB1, which is proposed to participate in cell signaling and defense against bacterium.[24]
Interactions
CHID1 is only strongly suggested to interact with one other protein. The transmembrane protein stabilin-1 has been detected as an interactant by in vitro, in vivo, and yeast two-hybrid assays.[10] STAB1 is a large transmembrane receptor protein which may function in many aspects such as lymphocyte homing or angiogenesis. It is expressed at over twice the average gene level and is expected to play a role in cell defense against bacterium.[25] This interaction may give insight towards the function of both proteins as a whole.
Clinical Significance
Overall, the significance of CHID1 is unknown. Many expression profiles of CHID1 show that its expression does not change in any common diseases or conditions as currently studied.[26] Given the lack of knowledge about the function of CHID1, it is difficult to further study its role within the human condition.
References
- GRCh38: Ensembl release 89: ENSG00000177830 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000025512 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Taylor TD, Noguchi H, Totoki Y, Toyoda A, Kuroki Y, Dewar K, Lloyd C, Itoh T, Takeda T, Kim DW, She X, Barlow KF, Bloom T, Bruford E, Chang JL, Cuomo CA, Eichler E, FitzGerald MG, Jaffe DB, LaButti K, Nicol R, Park HS, Seaman C, Sougnez C, Yang X, Zimmer AR, Zody MC, Birren BW, Nusbaum C, Fujiyama A, Hattori M, Rogers J, Lander ES, Sakaki Y (Mar 2006). "Human chromosome 11 DNA sequence and analysis including novel gene identification". Nature. 440 (7083): 497–500. Bibcode:2006Natur.440..497T. doi:10.1038/nature04632. PMID 16554811.
- "CHID1 Gene". GeneCards. Weizmann Institute of Science. Retrieved 30 April 2013.
- "Homo sapiens complex locus CHID1, encoding chitinase domain containing 1". AceView. NCBI. Retrieved 30 April 2013.
- GeneCards Human Gene Database. "CHID1 Gene - GeneCards | CHID1 Protein | CHID1 Antibody". GeneCards. Retrieved 2013-07-26.
- "Human chr11:867,357-915,058 - UCSC Genome Browser v286". Genome.ucsc.edu. Retrieved 2013-07-26.
- Kzhyshkowska J, Mamidi S, Gratchev A, Kremmer E, Schmuttermaier C, Krusell L, Haus G, Utikal J, Schledzewski K, Scholtze J, Goerdt S (Apr 2006). "Novel stabilin-1 interacting chitinase-like protein (SI-CLP) is up-regulated in alternatively activated macrophages and secreted via lysosomal pathway". Blood. 107 (8): 3221–8. doi:10.1182/blood-2005-07-2843. PMID 16357325.
- "HomoloGene Home". Ncbi.nlm.nih.gov. Retrieved 2013-07-26.
- "BLAST: Basic Local Alignment Search Tool". Blast.ncbi.nlm.nih.gov. 2013-06-20. Retrieved 2013-07-26.
- "Welcome to". Psort.org. 2010-05-17. Retrieved 2013-07-26.
- "LipoP 1.0 Server". Cbs.dtu.dk. Retrieved 2013-07-26.
- Meng G, Zhao Y, Bai X, Liu Y, Green TJ, Luo M, Zheng X (Dec 2010). "Structure of human stabilin-1 interacting chitinase-like protein (SI-CLP) reveals a saccharide-binding cleft with lower sugar-binding selectivity". The Journal of Biological Chemistry. 285 (51): 39898–904. doi:10.1074/jbc.M110.130781. PMC 3000971. PMID 20724479.
- "BioGPS - your Gene Portal System".
- Danielle Thierry-Mieg; Jean Thierry-Mieg, NCBI/NLM/NIH. "AceView: Gene:CHID1, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView". Ncbi.nlm.nih.gov. Retrieved 2013-07-26.
- GeneCards Human Gene Database. "CHID1 Gene - GeneCards | CHID1 Protein | CHID1 Antibody". GeneCards. Retrieved 2013-07-26.
- "Genomatix: Login Page". Genomatix.de. Retrieved 2013-07-26.
- "FlyBase Gene Report: Dmel\CG8460". Flybase.org. Retrieved 2013-07-26.
- "Experiment Detail :: Allen Brain Atlas: Mouse Brain". Mouse.brain-map.org. Retrieved 2013-07-26.
- "Experiment Detail :: Allen Brain Atlas: Mouse Brain". Mouse.brain-map.org. Retrieved 2013-07-26.
- Kzhyshkowska J, Mamidi S, Gratchev A, Kremmer E, Schmuttermaier C, Krusell L, Haus G, Utikal J, Schledzewski K, Scholtze J, Goerdt S (Apr 2006). "Novel stabilin-1 interacting chitinase-like protein (SI-CLP) is up-regulated in alternatively activated macrophages and secreted via lysosomal pathway". Blood. 107 (8): 3221–8. doi:10.1182/blood-2005-07-2843. PMID 16357325.
- Danielle Thierry-Mieg; Jean Thierry-Mieg, NCBI/NLM/NIH. "AceView: Gene:STAB1, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView". Ncbi.nlm.nih.gov. Retrieved 2013-07-26.
- Danielle Thierry-Mieg; Jean Thierry-Mieg, NCBI/NLM/NIH. "AceView: Gene:STAB1, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView". Ncbi.nlm.nih.gov. Retrieved 2013-07-26.
- "Home - GEO Profiles - NCBI". Ncbi.nlm.nih.gov. 2013-03-25. Retrieved 2013-07-26.



