Cyanobacterial morphology
Cyanobacterial morphology refers to the form or shape of cyanobacteria. Cyanobacteria are a large and diverse phylum of bacteria defined by their unique combination of pigments and their ability to perform oxygenic photosynthesis.[2][3]
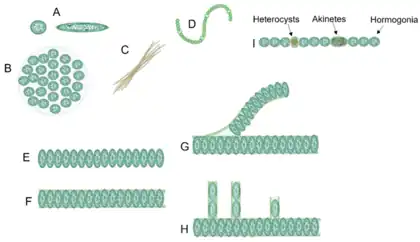
Cyanobacteria often live in colonial aggregates that can take a multitude of forms.[3] Of particular interest among the many species of cyanobacteria are those that live colonially in elongate hair-like structures, known as trichomes. These filamentous species can contain hundreds to thousands of cells.[3] They often dominate the upper layers of microbial mats found in extreme environments such as hot springs, hypersaline water, deserts and polar regions,[4] as well as being widely distributed in more mundane environments.[3]
Many filamentous species are also motile, gliding along their long axis, and displaying photomovement by which a trichome modulates its gliding according to the incident light. The latter has been found to play an important role in guiding the trichomes to optimal lighting conditions, which can either inhibit the cells if the incident light is too weak, or damage the cells if too strong.[3]
Overview
Cellular functions require a well-organized and coordinated internal structure to operate effectively. Cells need to build, sustain, and sometimes modify their shape, which allows them to rapidly change their behaviour in response to external factors. During different life cycle stages, such as cell growth, cell division or cell differentiation, internal structures must dynamically adapt to the current requirements. In eukaryotes, these manifold tasks are fulfilled by the cytoskeleton: proteinaceous polymers that assemble into stable or dynamic filaments or tubules in vivo and in vitro. The eukaryotic cytoskeleton is historically divided into three classes: the actin filaments (consisting of actin monomers), the microtubules (consisting of tubulin subunits) and the intermediate filaments (IFs), although other cytoskeletal classes have been identified in recent years.[5][6] Only the collaborative work of all three cytoskeletal systems enables proper cell mechanics.[7][8]
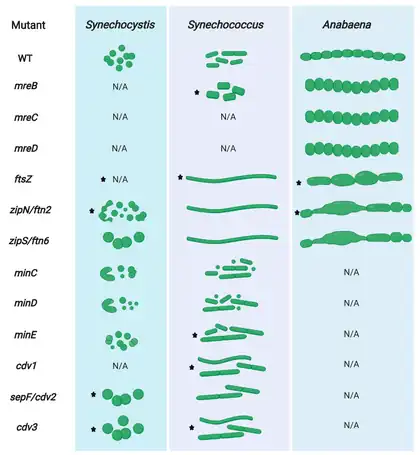
The long-lasting dogma that prokaryotes, based on their simple cell shapes, do not require cytoskeletal elements was finally abolished by the discovery of FtsZ, a prokaryotic tubulin homolog,[9][10][11] and MreB, a bacterial actin homolog.[12][13] These discoveries started an intense search for other cytoskeletal proteins in bacteria and archaea which finally led to the identification of bacterial IF-like proteins such as Crescentin from Caulobacter crescentus[14] and even bacterial-specific cytoskeletal protein classes, including bactofilins.[15] Constant influx of new findings finally established that numerous prokaryotic cellular functions, including cell division, cell elongation or bacterial microcompartment segregation are governed by the prokaryotic cytoskeleton.[16][17][8]
Cyanobacteria are today's only known prokaryotes capable of performing oxygenic photosynthesis. Based on the presence of an outer membrane, cyanobacteria are generally considered Gram-negative bacteria. However, unlike other Gram-negative bacteria, cyanobacteria contain an unusually thick peptidoglycan (PG) layer between the inner and outer membrane, thus containing features of both Gram phenotypes.[18][19][20] Additionally, the degree of PG crosslinking is much higher in cyanobacteria than in other Gram-negative bacteria, although teichoic acids, typically present in Gram-positive bacteria, are absent.[21][8]
While Cyanobacteria are monophyletic,[22] their cellular morphologies are extremely diverse and range from unicellular species to complex cell-differentiating, multicellular species. Based on this observation, cyanobacteria have been classically divided into five subsections.[23] Subsection I cyanobacteria (Chroococcales) are unicellular and divide by binary fission or budding, whereas subsection II cyanobacteria (Pleurocapsales) are also unicellular but can undergo multiple fission events, giving rise to many small daughter cells termed baeocytes. Subsection III comprises multicellular, non-cell differentiating cyanobacteria (Oscillatoriales) and subsection IV and V cyanobacteria (Nostocales and Stigonematales) are multicellular, cell differentiating cyanobacteria that form specialized cell types in the absence of combined nitrogen (heterocysts), during unfavorable conditions (akinetes) or to spread and initiate symbiosis (hormogonia). Whereas subsections III and IV form linear cell filaments (termed trichomes) that are surrounded by a common sheath, subsection V can produce lateral branches and/or divide in multiple planes, establishing multiseriate trichomes.[23] Considering this complex morphology, it was postulated that certain subsection V-specific (cytoskeletal) proteins could be responsible for this phenotype. However, no specific gene was identified whose distribution was specifically correlated with the cell morphology among different cyanobacterial subsections.[24][25] Therefore, it seems more likely that differential expression of cell growth and division genes rather than the presence or absence of a single gene is responsible for the cyanobacterial morphological diversity.[24][26][8]
Morphogenesis
Morphogenesis is the biological process that causes an organism to develop its shape. Cyanobacteria show a high degree of morphological diversity and can undergo a variety of cellular differentiation processes in order to adapt to certain environmental conditions. This helps them thrive in almost every habitat on Earth, ranging from freshwater to marine and terrestrial habitats, including even symbiotic interactions.[27] [8]
One factor which can drive morphological changes in cyanobacteria is light. As cyanobacteria are bacteria that use light to fuel their energy-producing photosynthetic machinery they depend on perceiving light in order to optimize their response and to avoid harmful light that could result in the formation of reactive oxygen species and subsequently in their death.[28] Optimal light conditions may be defined by quantity (irradiance), duration (day-night cycle) and wavelength (color of light). The photosynthetically usable light range of the solar spectrum is generally referred to as PAR (photosynthetically active radiation), but some cyanobacteria may expand on PAR by not only absorbing in the visible spectrum, but also the near-infrared light spectrum. This employs a variety of chlorophylls and allows phototrophic growth up to a wavelength of 750 nm.[29] To sense the light across this range of wavelengths, cyanobacteria possess various photoreceptors of the phytochrome superfamily.[30][8]
Morphological plasticity, or the ability of one cell to alternate between different shapes, is a common strategy of many bacteria in response to environmental changes or as part of their normal life cycle.[31][32][33] Bacteria may alter their shape by simpler transitions from rod to coccoid (and vice versa) as in Escherichia coli,[34] by more complex transitions while establishing multicellularity[31] or by the development of specialized cells, structures or appendages where the population presents a pleomorphic lifestyle.[35] The precise molecular circuits that govern those morphological changes are yet to be identified, however, a so-far constant factor is that the cell shape is determined by the rigid PG sacculus which consists of glycan strands crosslinked by peptides. To grow, cells must synthesize new PG while breaking down the existent polymer to insert the newly synthesized material. How cells grow and elongate has been extensively reviewed in model organisms of both, rod-shaped[36][37] and coccoid bacteria.[38] The molecular basis for morphological plasticity and pleomorphism in more complex bacteria, however, is slowly being elucidated as well.[33][8]
Despite their morphological complexity, cyanobacteria contain all conserved and so far known bacterial morphogens.[8] Understanding cyanobacterial morphogenesis is challenging, as there are numerous morphotypes among cyanobacterial taxa, which can also vary within a given strain during its life cycle.[23] Changes in cellular or even trichome morphologies are tasks that would require active cell wall remodelling and thus far no genes attributed to the different morphotypes have been identified in cyanobacteria.[24] Therefore, the most likely scenario is that genes or their products are differentially regulated during these cell morphology transitions,[26] as it has been hypothesized for most bacteria.[33] In multicellular cyanobacteria, division of labor between cells within a trichome is achieved by different cell programing strategies. Thus, gene regulation occurs differentially in these specific cell types [30,97,98].[8]
Diversity of forms
Cyanobacteria present remarkable variability in terms of morphology: from unicellular and colonial to multicellular filamentous forms. Their cell size varies from less than 1 µm in diameter (picocyanobacteria) up to 100 µm (some tropical forms in the genus Oscillatoria)[39][40][41]
Filamentous forms exhibit functional cell differentiation such as heterocysts (for nitrogen fixation), akinetes (resting stage cells), and hormogonia (reproductive, motile filaments). These, together with the intercellular connections they possess, are considered the first signs of multicellularity.[31][42][43][44]
Many cyanobacteria form motile filaments of cells, called hormogonia, that travel away from the main biomass to bud and form new colonies elsewhere.[45][46] The cells in a hormogonium are often thinner than in the vegetative state, and the cells on either end of the motile chain may be tapered. To break away from the parent colony, a hormogonium often must tear apart a weaker cell in a filament, called a necridium.
scale bars about 10 µm

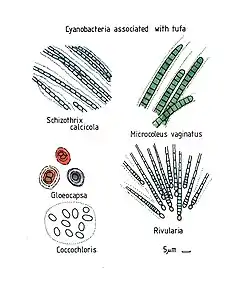
Schizothrix calcicola, Gloeocapsa, Coccochloris, Microcoleus vaginatus and Rivularia
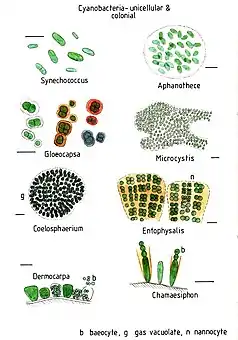
Colonial and unicellular
In aquatic habitats, unicellular cyanobacteria are considered as an important group regarding abundance, diversity, and ecological character.[47] Unicellular cyanobacteria have spherical, ovoid, or cylindrical cells that may aggregate into irregular or regular colonies bound together by the mucous matrix (mucilage) secreted during the growth of the colony.[48] Based on the species, the number of cells in each colony may vary from two to several thousand.[47][1]
Each individual cell (each single cyanobacterium) typically has a thick, gelatinous cell wall.[49] They lack flagella, but hormogonia of some species can move about by gliding along surfaces.[50]
 Merismopedia forms rectangular colonies held together by a mucilaginous matrix. Species in this genus divide in only two directions, creating a characteristic grid-like pattern arranged in rows and flats.[51]
Merismopedia forms rectangular colonies held together by a mucilaginous matrix. Species in this genus divide in only two directions, creating a characteristic grid-like pattern arranged in rows and flats.[51]
 Colonies of Nostoc pruniforme "jelly balls"
Colonies of Nostoc pruniforme "jelly balls" Colonial cyanobacteria Stratonostoc
Colonial cyanobacteria Stratonostoc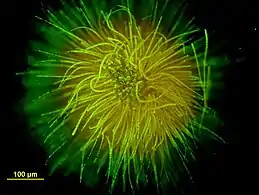 Ball-shaped colony of Gloeotrichia echinulata
Ball-shaped colony of Gloeotrichia echinulata Cyanobacterial colony of Lyngbya majuscula
Cyanobacterial colony of Lyngbya majuscula
Filamentous and multicellular
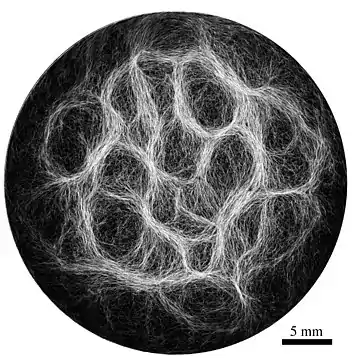 Example of filamentous cyanobacteria structure (Oscillatoria lutea) showing a reticulate pattern [52]
Example of filamentous cyanobacteria structure (Oscillatoria lutea) showing a reticulate pattern [52]
Some filamentous species can differentiate into several different cell types:
- vegetative cells – the normal, photosynthetic cells that are formed under favorable growing conditions
- akinetes – climate-resistant spores that may form when environmental conditions become harsh
- thick-walled heterocysts – which contain the enzyme nitrogenase vital for nitrogen fixation[53][54][55] in an anaerobic environment due to its sensitivity to oxygen.[55]
Many of the multicellular filamentous forms of Oscillatoria are capable of a waving motion; the filament oscillates back and forth. In water columns, some cyanobacteria float by forming gas vesicles, as in archaea.[56] These vesicles are not organelles as such. They are not bounded by lipid membranes but by a protein sheath.

A–C: Microcoleus steenstrupii D–E: Tolypothrix desertorum F: Scytonema cf. calcicola G: S. cf. calcicola H: S. cf. c alcicola
Scale bar =10 µm
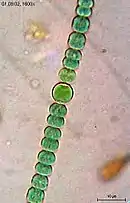

 Helical filaments of cyanobacteria
Helical filaments of cyanobacteria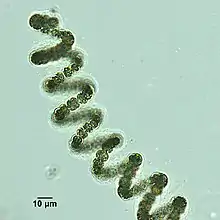 Helical filament from Dolichospermum
Helical filament from Dolichospermum Lyngbya species form long, unbranching filaments inside rigid mucilaginous sheaths which can form tangles or mats, intermixed with other phytoplankton species
Lyngbya species form long, unbranching filaments inside rigid mucilaginous sheaths which can form tangles or mats, intermixed with other phytoplankton species
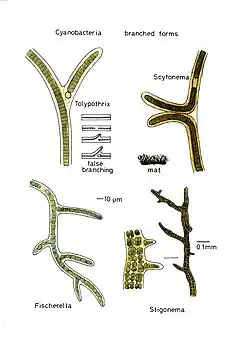
Branched
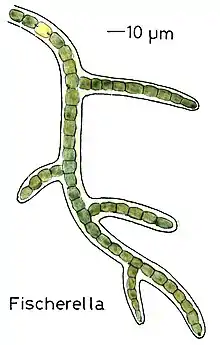
 True branching phenotype of a Fischerella thermalis colony
True branching phenotype of a Fischerella thermalis colony
Heterocysts
Heterocysts are specialized nitrogen-fixing cells formed during nitrogen starvation by some filamentous cyanobacteria, such as Nostoc punctiforme, Cylindrospermum stagnale, and Anabaena sphaerica.[58] They fix nitrogen from atmospheric N2 using the enzyme nitrogenase, in order to provide the cells in the filament with nitrogen for biosynthesis.[59]
Movement
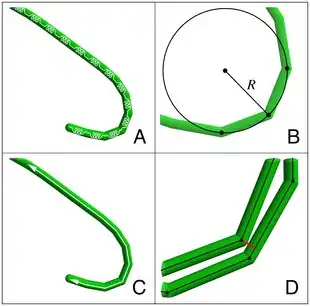
Cyanobacteria are ubiquitous, finding habitats in most water bodies and in extreme environments such as the polar regions, deserts, brine lakes and hot springs.[60][61][62] They have also evolved surprisingly complex collective behaviours that lie at the boundary between single-celled and multicellular life. For example, filamentous cyanobacteria live in long chains of cells that bundle together into larger structures including biofilms, biomats and stromatolites.[63][64] These large colonies provide a rigid, stable and long-term environment for their communities of bacteria. In addition, cyanobacteria-based biofilms can be used as bioreactors to produce a wide range of chemicals, including biofuels like biodiesel and ethanol.[65] However, despite their importance to the history of life on Earth, and their commercial and environmental potentials, there remain basic questions of how filamentous cyanobacteria move, respond to their environment and self-organize into collective patterns and structures.[52]
All known cyanobacteria lack flagella;[66] however, many filamentous species move on surfaces by gliding, a form of locomotion where no physical appendages are seen to aid movement.[67] The actual mechanism behind gliding is not fully understood, although over a century has elapsed since its discovery.[68][69] One theory suggests that gliding motion in cyanobacteria is mediated by the continuous secretion of polysaccharides through pores on individual cells.[70][71][72] Another theory suggests that gliding motion involves the use of type IV pili, polymeric assemblies of the protein pilin,[73] as the driving engines of motion.[74][75][76] However, it is not clear how the action of these pili would lead to motion, with some suggesting they retract,[77] while others suggest they push,[76] to generate forces. Other scholars have suggested surface waves generated by the contraction of a fibril layer as the mechanism behind gliding motion in Oscillatoria.[78][79] Recent work also suggests that shape fluctuations and capillary forces could be involved in gliding motion.[80][52]
Through collective interaction, filamentous cyanobacteria self-organize into colonies or biofilms, symbiotic communities found in a wide variety of ecological niches. Their larger-scale collective structures are characterized by diverse shapes including bundles, vortices and reticulate patterns.[81][82] Similar patterns have been observed in fossil records.[83][82][84] For filamentous cyanobacteria, the mechanics of the filaments is known to contribute to self-organization, for example in determining how one filament will bend when in contact with other filaments or obstacles.[85] Further, biofilms and biomats show some remarkably conserved macro-mechanical properties, typically behaving as viscoelastic materials with a relaxation time of about 20 min.[86][52]
Cyanobacteria have strict light requirements. Too little light can result in insufficient energy production, and in some species may cause the cells to resort to heterotrophic respiration.[4] Too much light can inhibit the cells, decrease photosynthesis efficiency and cause damage by bleaching. UV radiation is especially deadly for cyanobacteria, with normal solar levels being significantly detrimental for these microorganisms in some cases.[87][88][3]
Filamentous cyanobacteria that live in microbial mats often migrate vertically and horizontally within the mat in order to find an optimal niche that balances their light requirements for photosynthesis against their sensitivity to photodamage. For example, the filamentous cyanobacteria Oscillatoria sp. and Spirulina subsalsa found in the hypersaline benthic mats of Guerrero Negro, Mexico migrate downwards into the lower layers during the day in order to escape the intense sunlight and then rise to the surface at dusk.[89] In contrast, the population of Microcoleus chthonoplastes found in hypersaline mats at Salin-de-Giraud, Camargue, France migrate to the upper layer of the mat during the day and are spread homogenously through the mat at night.[90] An in vitro experiment using P. uncinatum also demonstrated this species' tendency to migrate in order to avoid damaging radiation.[87][88] These migrations are usually the result of some sort of photomovement, although other forms of taxis can also play a role.[91][3]
Many species of cyanobacteria are capable of gliding. Gliding is a form of cell movement that differs from crawling or swimming in that it does not rely on any obvious external organ or change in cell shape and it occurs only in the presence of a substrate.[92][93] Gliding in filamentous cyanobacteria appears to be powered by a "slime jet" mechanism, in which the cells extrude a gel that expands quickly as it hydrates providing a propulsion force,[94][95] although some unicellular cyanobacteria use type IV pili for gliding.[96] Individual cells in a trichome have two sets of pores for extruding slime. Each set is organized in a ring at the cell septae and extrudes slime at an acute angle.[97] The sets extrude slime in opposite directions and so only one set is likely to be activated during gliding. An alternative hypothesis is that the cells use contractive elements that produce undulations running over the surface inside the slime tube like an earthworm.[98] The trichomes rotate in a spiral fashion, the angle of which corresponds with the pitch angle of Castenholz's contractile trichomes.[3]


The cells appear to coordinate their gliding direction by an electrical potential that establishes polarity in the trichomes, and thus establishes a "head" and the "tail".[99] Trichomes usually reverse their polarity randomly with an average period on the order of minutes to hours.[100][101] Many species also form a semi-rigid sheath that is left behind as a hollow tube as the trichome moves forward. When the trichome reverses direction, it can move back into the sheath or break out.[102][3]
Oscillatoria is a genus of filamentous cyanobacterium named after the oscillation in its movement. Filaments in colonies slide back and forth against each other until the whole mass is reoriented to its light source. Oscillatoria is mainly blue-green or brown-green and is commonly found in watering-troughs. It reproduces by fragmentation forming long filaments of cells which can break into fragments called hormogonia. The hormogonia can then grow into new, longer filaments.
Häder's cyanograph experiment

In 1987, Häder demonstrated that trichomes can position themselves quite precisely within their environment through photomovement. In Häder's cyanograph experiment a photographic negative is projected onto a Petri dish containing a culture of Phormidium uncinatum.[103][104] After a few hours, the trichomes move away from the darker areas onto the lighter areas, forming a photographic positive on the culture. The experiment demonstrates that photomovement is effective not just for discrete light traps, but for minutely patterned, continuously differentiated light fields as well.[3]
References
- Mehdizadeh Allaf, Malihe; Peerhossaini, Hassan (2022-03-24). "Cyanobacteria: Model Microorganisms and Beyond". Microorganisms. MDPI AG. 10 (4): 696. doi:10.3390/microorganisms10040696. ISSN 2076-2607. PMC 9025173. PMID 35456747.
 Modified material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Whitton, Brian A.; Potts, Malcolm (2012). "Introduction to the Cyanobacteria". Ecology of Cyanobacteria II. pp. 1–13. doi:10.1007/978-94-007-3855-3_1. ISBN 978-94-007-3854-6.
- Tamulonis, Carlos; Postma, Marten; Kaandorp, Jaap (2011). "Modeling Filamentous Cyanobacteria Reveals the Advantages of Long and Fast Trichomes for Optimizing Light Exposure". PLOS ONE. 6 (7): e22084. Bibcode:2011PLoSO...622084T. doi:10.1371/journal.pone.0022084. PMC 3138769. PMID 21789215.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Stal, Lucas J. (5 July 2012). "Cyanobacterial Mats and Stromatolites". In Whitton, Brian A. (ed.). Ecology of Cyanobacteria II. pp. 65–126. ISBN 9789400738553.
- Moseley, James B. (2013). "An expanded view of the eukaryotic cytoskeleton". Molecular Biology of the Cell. American Society for Cell Biology (ASCB). 24 (11): 1615–1618. doi:10.1091/mbc.e12-10-0732. ISSN 1059-1524. PMC 3667716. PMID 23722945.
- Alberts, Bruce; Heald, Rebecca; Johnson, Alexander; Morgan, David Owen; Raff, Martin C.; Roberts, K.; Walter, Peter (2022). Molecular biology of the cell. New York, NY. ISBN 978-0-393-88482-1. OCLC 1276902141.
{{cite book}}: CS1 maint: location missing publisher (link) - Huber, Florian; Boire, Adeline; López, Magdalena Preciado; Koenderink, Gijsje H (2015). "Cytoskeletal crosstalk: when three different personalities team up". Current Opinion in Cell Biology. Elsevier BV. 32: 39–47. doi:10.1016/j.ceb.2014.10.005. ISSN 0955-0674. PMID 25460780. S2CID 40360166.
- Springstein, Benjamin L.; Nürnberg, Dennis J.; Weiss, Gregor L.; Pilhofer, Martin; Stucken, Karina (2020-12-17). "Structural Determinants and Their Role in Cyanobacterial Morphogenesis". Life. MDPI AG. 10 (12): 355. Bibcode:2020Life...10..355S. doi:10.3390/life10120355. ISSN 2075-1729. PMC 7766704. PMID 33348886. Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- Bi, Erfei; Lutkenhaus, Joe (1991). "FtsZ ring structure associated with division in Escherichia coli". Nature. Springer Science and Business Media LLC. 354 (6349): 161–164. Bibcode:1991Natur.354..161B. doi:10.1038/354161a0. ISSN 0028-0836. PMID 1944597. S2CID 4329947.
- de Boer, Piet; Crossley, Robin; Rothfield, Lawrence (1992). "The essential bacterial cell-division protein FtsZ is a GTPase". Nature. Springer Science and Business Media LLC. 359 (6392): 254–256. Bibcode:1992Natur.359..254D. doi:10.1038/359254a0. ISSN 0028-0836. PMID 1528268. S2CID 2748757.
- Löwe, Jan; Amos, Linda A. (1998). "Crystal structure of the bacterial cell-division protein FtsZ". Nature. Springer Science and Business Media LLC. 391 (6663): 203–206. Bibcode:1998Natur.391..203L. doi:10.1038/34472. ISSN 0028-0836. PMID 9428770. S2CID 4330857.
- Bork, P; Sander, C; Valencia, A (1992-08-15). "An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins". Proceedings of the National Academy of Sciences. 89 (16): 7290–7294. Bibcode:1992PNAS...89.7290B. doi:10.1073/pnas.89.16.7290. ISSN 0027-8424. PMC 49695. PMID 1323828.
- van den Ent, Fusinita; Amos, Linda A.; Löwe, Jan (2001). "Prokaryotic origin of the actin cytoskeleton". Nature. Springer Science and Business Media LLC. 413 (6851): 39–44. Bibcode:2001Natur.413...39V. doi:10.1038/35092500. ISSN 0028-0836. PMID 11544518. S2CID 4427828.
- Ausmees, Nora; Kuhn, Jeffrey R; Jacobs-Wagner, Christine (2003). "The Bacterial Cytoskeleton". Cell. Elsevier BV. 115 (6): 705–713. doi:10.1016/s0092-8674(03)00935-8. ISSN 0092-8674. PMID 14675535. S2CID 14459851.
- Kühn, Juliane; Briegel, Ariane; Mörschel, Erhard; Kahnt, Jörg; Leser, Katja; Wick, Stephanie; Jensen, Grant J; Thanbichler, Martin (2009-12-03). "Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus". The EMBO Journal. Wiley. 29 (2): 327–339. doi:10.1038/emboj.2009.358. ISSN 0261-4189. PMC 2824468. PMID 19959992.
- Lin, Lin; Thanbichler, Martin (2013). "Nucleotide-independent cytoskeletal scaffolds in bacteria". Cytoskeleton. Wiley. 70 (8): 409–423. doi:10.1002/cm.21126. ISSN 1949-3584. PMID 23852773. S2CID 40504066.
- Wagstaff, James; Löwe, Jan (2018-01-22). "Prokaryotic cytoskeletons: protein filaments organizing small cells". Nature Reviews Microbiology. Springer Science and Business Media LLC. 16 (4): 187–201. doi:10.1038/nrmicro.2017.153. ISSN 1740-1526. PMID 29355854. S2CID 3537215.
- Videau, Patrick; Rivers, Orion S.; Ushijima, Blake; Oshiro, Reid T.; Kim, Min Joo; Philmus, Benjamin; Cozy, Loralyn M. (2016-04-15). "Mutation of the murC and murB Genes Impairs Heterocyst Differentiation in Anabaena sp. Strain PCC 7120". Journal of Bacteriology. American Society for Microbiology. 198 (8): 1196–1206. doi:10.1128/jb.01027-15. ISSN 0021-9193. PMC 4859589. PMID 26811320.
- Hoiczyk, E; Baumeister, W (1995). "Envelope structure of four gliding filamentous cyanobacteria". Journal of Bacteriology. American Society for Microbiology. 177 (9): 2387–2395. doi:10.1128/jb.177.9.2387-2395.1995. ISSN 0021-9193. PMC 176896. PMID 7730269.
- Gumbart, James C.; Beeby, Morgan; Jensen, Grant J.; Roux, Benoît (2014-02-20). "Escherichia coli Peptidoglycan Structure and Mechanics as Predicted by Atomic-Scale Simulations". PLOS Computational Biology. Public Library of Science (PLoS). 10 (2): e1003475. Bibcode:2014PLSCB..10E3475G. doi:10.1371/journal.pcbi.1003475. ISSN 1553-7358. PMC 3930494. PMID 24586129.
- Hoiczyk, Egbert; Hansel, Alfred (2000). "Cyanobacterial Cell Walls: News from an Unusual Prokaryotic Envelope". Journal of Bacteriology. American Society for Microbiology. 182 (5): 1191–1199. doi:10.1128/jb.182.5.1191-1199.2000. ISSN 0021-9193. PMC 94402. PMID 10671437.
- Schirrmeister, Bettina E; Antonelli, Alexandre; Bagheri, Homayoun C (2011-02-14). "The origin of multicellularity in cyanobacteria". BMC Evolutionary Biology. Springer Science and Business Media LLC. 11 (1): 45. doi:10.1186/1471-2148-11-45. ISSN 1471-2148. PMC 3271361. PMID 21320320.
- Rippka, Rosmarie; Stanier, Roger Y.; Deruelles, Josette; Herdman, Michael; Waterbury, John B. (1979-03-01). "Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria". Microbiology. Microbiology Society. 111 (1): 1–61. doi:10.1099/00221287-111-1-1. ISSN 1350-0872.
- Dagan, Tal; Roettger, Mayo; Stucken, Karina; Landan, Giddy; Koch, Robin; Major, Peter; Gould, Sven B.; Goremykin, Vadim V.; Rippka, Rosmarie; Tandeau de Marsac, Nicole; Gugger, Muriel; Lockhart, Peter J.; Allen, John F.; Brune, Iris; Maus, Irena; Pühler, Alfred; Martin, William F. (2012-12-07). "Genomes of Stigonematalean Cyanobacteria (Subsection V) and the Evolution of Oxygenic Photosynthesis from Prokaryotes to Plastids". Genome Biology and Evolution. Oxford University Press (OUP). 5 (1): 31–44. doi:10.1093/gbe/evs117. ISSN 1759-6653. PMC 3595030. PMID 23221676.
- Shih, Patrick M.; Wu, Dongying; Latifi, Amel; Axen, Seth D.; Fewer, David P.; Talla, Emmanuel; Calteau, Alexandra; Cai, Fei; Tandeau de Marsac, Nicole; Rippka, Rosmarie; Herdman, Michael; Sivonen, Kaarina; Coursin, Therese; Laurent, Thierry; Goodwin, Lynne; Nolan, Matt; Davenport, Karen W.; Han, Cliff S.; Rubin, Edward M.; Eisen, Jonathan A.; Woyke, Tanja; Gugger, Muriel; Kerfeld, Cheryl A. (2012-12-31). "Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing". Proceedings of the National Academy of Sciences. 110 (3): 1053–1058. doi:10.1073/pnas.1217107110. ISSN 0027-8424. PMC 3549136. PMID 23277585.
- Koch, Robin; Kupczok, Anne; Stucken, Karina; Ilhan, Judith; Hammerschmidt, Katrin; Dagan, Tal (2017-08-31). "Plasticity first: molecular signatures of a complex morphological trait in filamentous cyanobacteria". BMC Evolutionary Biology. Springer Science and Business Media LLC. 17 (1): 209. doi:10.1186/s12862-017-1053-5. ISSN 1471-2148. PMC 5580265. PMID 28859625.
- Gaysina, Lira A.; Saraf, Aniket; Singh, Prashant (2019). "Cyanobacteria in Diverse Habitats". Cyanobacteria. Elsevier. pp. 1–28. doi:10.1016/b978-0-12-814667-5.00001-5. ISBN 9780128146675. S2CID 135429562.
- Pospíšil, Pavel (2016-12-26). "Production of Reactive Oxygen Species by Photosystem II as a Response to Light and Temperature Stress". Frontiers in Plant Science. Frontiers Media SA. 7: 1950. doi:10.3389/fpls.2016.01950. ISSN 1664-462X. PMC 5183610. PMID 28082998.
- Larkum, A. W. D.; Ritchie, R. J.; Raven, J. A. (2018). "Living off the Sun: Chlorophylls, bacteriochlorophylls and rhodopsins". Photosynthetica. 56: 11–43. doi:10.1007/s11099-018-0792-x. S2CID 4907693.
- Wiltbank, Lisa B.; Kehoe, David M. (2018-11-08). "Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors". Nature Reviews Microbiology. Springer Science and Business Media LLC. 17 (1): 37–50. doi:10.1038/s41579-018-0110-4. ISSN 1740-1526. PMID 30410070. S2CID 256744429.
- Claessen, Dennis; Rozen, Daniel E.; Kuipers, Oscar P.; Søgaard-Andersen, Lotte; Van Wezel, Gilles P. (2014). "Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies". Nature Reviews Microbiology. 12 (2): 115–124. doi:10.1038/nrmicro3178. hdl:11370/0db66a9c-72ef-4e11-a75d-9d1e5827573d. PMID 24384602. S2CID 20154495.
- Zhu, Zaichun; Piao, Shilong; Myneni, Ranga B.; Huang, Mengtian; Zeng, Zhenzhong; Canadell, Josep G.; Ciais, Philippe; Sitch, Stephen; Friedlingstein, Pierre; Arneth, Almut; Cao, Chunxiang; Cheng, Lei; Kato, Etsushi; Koven, Charles; Li, Yue; Lian, Xu; Liu, Yongwen; Liu, Ronggao; Mao, Jiafu; Pan, Yaozhong; Peng, Shushi; Peñuelas, Josep; Poulter, Benjamin; Pugh, Thomas A. M.; Stocker, Benjamin D.; Viovy, Nicolas; Wang, Xuhui; Wang, Yingping; Xiao, Zhiqiang; Yang, Hui; Zaehle, Sönke; Zeng, Ning (2016-04-25). "Greening of the Earth and its drivers". Nature Climate Change. Springer Science and Business Media LLC. 6 (8): 791–795. Bibcode:2016NatCC...6..791Z. doi:10.1038/nclimate3004. ISSN 1758-678X. S2CID 7980894.
- Caccamo, Paul D.; Brun, Yves V. (2018). "The Molecular Basis of Noncanonical Bacterial Morphology". Trends in Microbiology. Elsevier BV. 26 (3): 191–208. doi:10.1016/j.tim.2017.09.012. ISSN 0966-842X. PMC 5834356. PMID 29056293.
- Lange, R; Hengge-Aronis, R (1991). "Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S". Journal of Bacteriology. American Society for Microbiology. 173 (14): 4474–4481. doi:10.1128/jb.173.14.4474-4481.1991. ISSN 0021-9193. PMC 208111. PMID 1648559.
- Young, Kevin D. (2006). "The Selective Value of Bacterial Shape". Microbiology and Molecular Biology Reviews. American Society for Microbiology. 70 (3): 660–703. doi:10.1128/mmbr.00001-06. ISSN 1092-2172. PMC 1594593. PMID 16959965.
- Typas, Athanasios; Banzhaf, Manuel; Gross, Carol A.; Vollmer, Waldemar (2011-12-28). "From the regulation of peptidoglycan synthesis to bacterial growth and morphology". Nature Reviews Microbiology. Springer Science and Business Media LLC. 10 (2): 123–136. doi:10.1038/nrmicro2677. ISSN 1740-1526. PMC 5433867. PMID 22203377.
- Egan, Alexander J. F.; Errington, Jeff; Vollmer, Waldemar (2020-05-18). "Regulation of peptidoglycan synthesis and remodelling". Nature Reviews Microbiology. Springer Science and Business Media LLC. 18 (8): 446–460. doi:10.1038/s41579-020-0366-3. ISSN 1740-1526. PMID 32424210. S2CID 256745837.
- Pinho, Mariana G.; Kjos, Morten; Veening, Jan-Willem (2013-08-16). "How to get (a)round: mechanisms controlling growth and division of coccoid bacteria" (PDF). Nature Reviews Microbiology. Springer Science and Business Media LLC. 11 (9): 601–614. doi:10.1038/nrmicro3088. ISSN 1740-1526. PMID 23949602. S2CID 205498610.
- Whitton, Brian A. (1992). "Diversity, Ecology, and Taxonomy of the Cyanobacteria". Photosynthetic Prokaryotes. Boston, MA: Springer US. pp. 1–51. doi:10.1007/978-1-4757-1332-9_1. ISBN 978-1-4757-1334-3.
- Schulz‐Vogt, Heide N; Angert, Esther R; Garcia‐Pichel, Ferran (2007-09-28), "Giant Bacteria", eLS, Wiley, doi:10.1002/9780470015902.a0020371, ISBN 9780470016176
- Jasser, Iwona; Callieri, Cristiana (2017-02-11). "Picocyanobacteria". Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis. Chichester, UK: John Wiley & Sons, Ltd. pp. 19–27. doi:10.1002/9781119068761.ch3. ISBN 9781119068761.
- Nürnberg, Dennis J.; Mariscal, Vicente; Parker, Jamie; Mastroianni, Giulia; Flores, Enrique; Mullineaux, Conrad W. (2014). "Branching and intercellular communication in the Section V cyanobacterium Mastigocladus laminosus, a complex multicellular prokaryote". Molecular Microbiology. 91 (5): 935–949. doi:10.1111/mmi.12506. hdl:10261/99110. PMID 24383541. S2CID 25479970.
- Herrero, Antonia; Stavans, Joel; Flores, Enrique (2016). "The multicellular nature of filamentous heterocyst-forming cyanobacteria". FEMS Microbiology Reviews. 40 (6): 831–854. doi:10.1093/femsre/fuw029. hdl:10261/140753. PMID 28204529.
- Aguilera, Anabella; Klemenčič, Marina; Sueldo, Daniela J.; Rzymski, Piotr; Giannuzzi, Leda; Martin, María Victoria (2021). "Cell Death in Cyanobacteria: Current Understanding and Recommendations for a Consensus on Its Nomenclature". Frontiers in Microbiology. 12: 631654. doi:10.3389/fmicb.2021.631654. PMC 7965980. PMID 33746925.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Risser DD, Chew WG, Meeks JC (April 2014). "Genetic characterization of the hmp locus, a chemotaxis-like gene cluster that regulates hormogonium development and motility in Nostoc punctiforme". Molecular Microbiology. 92 (2): 222–33. doi:10.1111/mmi.12552. PMID 24533832. S2CID 37479716.
- Khayatan B, Bains DK, Cheng MH, Cho YW, Huynh J, Kim R, Omoruyi OH, Pantoja AP, Park JS, Peng JK, Splitt SD, Tian MY, Risser DD (May 2017). "A Putative O-Linked β-N-Acetylglucosamine Transferase Is Essential for Hormogonium Development and Motility in the Filamentous Cyanobacterium Nostoc punctiforme". Journal of Bacteriology. 199 (9): e00075–17. doi:10.1128/JB.00075-17. PMC 5388816. PMID 28242721.
- Dvořák, Petr; Casamatta, Dale A.; Hašler, Petr; Jahodářová, Eva; Norwich, Alyson R.; Poulíčková, Aloisie (2017). "Diversity of the Cyanobacteria". Modern Topics in the Phototrophic Prokaryotes. Cham: Springer International Publishing. pp. 3–46. doi:10.1007/978-3-319-46261-5_1. ISBN 978-3-319-46259-2.
- Chorus, Ingrid; Bartram, Jamie (1999). Toxic cyanobacteria in water : a guide to their public health consequences, monitoring, and management. London: E & FN Spon. ISBN 0-419-23930-8. OCLC 40395794.
- Singh. Text Book of Botany Diversity of Microbes And Cryptogams. Rastogi Publications. ISBN 978-8171338894.
- "Differences between Bacteria and Cyanobacteria". Microbiology Notes. 2015-10-29. Retrieved 2018-01-21.
- Palinska, Katarzyna A.; Liesack, Werner; Rhiel, Erhard; Krumbein, W. E. (17 October 1996). "Phenotype variability of identical genotypes: the need for a combined approach in cyanobacterial taxonomy demonstrated on Merismopedia -like isolates". Archives of Microbiology. 166 (4): 224–233. doi:10.1007/s002030050378. PMID 8824145. S2CID 3022844.
- Faluweki, Mixon K.; Goehring, Lucas (2022). "Structural mechanics of filamentous cyanobacteria". Journal of the Royal Society Interface. The Royal Society. 19 (192). doi:10.1098/rsif.2022.0268. ISSN 1742-5662. PMC 9326267. PMID 35892203.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Meeks JC, Elhai J, Thiel T, Potts M, Larimer F, Lamerdin J, Predki P, Atlas R (2001). "An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium". Photosynthesis Research. 70 (1): 85–106. doi:10.1023/A:1013840025518. PMID 16228364. S2CID 8752382.
- Golden JW, Yoon HS (December 1998). "Heterocyst formation in Anabaena". Current Opinion in Microbiology. 1 (6): 623–9. doi:10.1016/s1369-5274(98)80106-9. PMID 10066546.
- Fay P (June 1992). "Oxygen relations of nitrogen fixation in cyanobacteria". Microbiological Reviews. 56 (2): 340–73. doi:10.1128/MMBR.56.2.340-373.1992. PMC 372871. PMID 1620069.
- Walsby AE (March 1994). "Gas vesicles". Microbiological Reviews. 58 (1): 94–144. doi:10.1128/MMBR.58.1.94-144.1994. PMC 372955. PMID 8177173.
- Schapiro, Igor (May 2014). "Ultrafast photochemistry of Anabaena Sensory Rhodopsin: Experiment and theory". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1837 (5): 589–597. doi:10.1016/j.bbabio.2013.09.014. PMID 24099700.
- Basic Biology (18 March 2016). "Bacteria".
- Wolk, C. Peter; Ernst, Annaliese; Elhai, Jeff (1994). "Heterocyst Metabolism and Development". In Donald A. Bryant (ed.). The Molecular Biology of Cyanobacteria. Advances in Photosynthesis and Respiration. pp. 769–823. doi:10.1007/978-94-011-0227-8_27. ISBN 978-0-7923-3273-2.
- Walter, M.R.; Bauld, J.; Brock, T.D. (1976). "Chapter 6.2 Microbiology and Morphogenesis of Columnar Stromatolites (Conophyton, Vacerrilla) from Hot Springs in Yellowstone National Park". Stromatolites. Developments in Sedimentology. Vol. 20. pp. 273–310. doi:10.1016/S0070-4571(08)71140-3. ISBN 9780444413765.
- Jones, B.; Renaut, R. W.; Rosen, M. R.; Ansdell, K. M. (2002). "Coniform Stromatolites from Geothermal Systems, North Island, New Zealand". PALAIOS. 17 (1): 84. Bibcode:2002Palai..17...84J. doi:10.1669/0883-1351(2002)017<0084:CSFGSN>2.0.CO;2. ISSN 0883-1351. S2CID 130120737.
- Wharton, Robert A.; Parker, Bruce C.; Simmons, George M. (1983). "Distribution, species composition and morphology of algal mats in Antarctic dry valley lakes". Phycologia. 22 (4): 355–365. doi:10.2216/i0031-8884-22-4-355.1.
- Whitton, Brian A, ed. (2012). Ecology of Cyanobacteria II. doi:10.1007/978-94-007-3855-3. ISBN 978-94-007-3854-6. S2CID 46736903.
- Stal, Lucas J. (2012). "Cyanobacterial Mats and Stromatolites". Ecology of Cyanobacteria II. pp. 65–125. doi:10.1007/978-94-007-3855-3_4. ISBN 978-94-007-3854-6.
- Farrokh, Parisa; Sheikhpour, Mojgan; Kasaeian, Alibakhsh; Asadi, Hassan; Bavandi, Roya (2019). "Cyanobacteria as an eco‐friendly resource for biofuel production: A critical review". Biotechnology Progress. 35 (5): e2835. doi:10.1002/btpr.2835. PMID 31063628. S2CID 147705730.
- Rippka, Rosmarie; Stanier, Roger Y.; Deruelles, Josette; Herdman, Michael; Waterbury, John B. (1979). "Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria". Microbiology. 111: 1–61. doi:10.1099/00221287-111-1-1.
- Hoiczyk, E. (2000). "Gliding motility in cyanobacteria: Observations and possible explanations". Archives of Microbiology. 174 (1–2): 11–17. doi:10.1007/s002030000187. PMID 10985737. S2CID 9927312.
- Hansgirg A. (1883) "Bemerkungen über die Bewegungen der Oscillarien". Bot. Ztg., 41: 831.
- Drews G. (1959) "Beitröge zur Kenntnis der phototaktischen Reaktionen der Cyanophyceen". Arch. Protistenk. 104: 389–430.
- Hosoi, Akimitsu (1951). "Secretion of the slime substance in Oscillatoria in relation to its movement". Shokubutsugaku Zasshi. 64 (751–752): 14–17. doi:10.15281/jplantres1887.64.14.
- Walsby, A. E. (1968). "Mucilage secretion and the movements of blue-green algae". Protoplasma. 65 (1–2): 223–238. doi:10.1007/BF01666380. S2CID 20310025.
- Hoiczyk, Egbert; Baumeister, Wolfgang (1998). "The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria". Current Biology. 8 (21): 1161–1168. doi:10.1016/S0960-9822(07)00487-3. PMID 9799733. S2CID 14384308.
- Craig, Lisa; Pique, Michael E.; Tainer, John A. (2004). "Type IV pilus structure and bacterial pathogenicity". Nature Reviews Microbiology. 2 (5): 363–378. doi:10.1038/nrmicro885. PMID 15100690. S2CID 10654430.
- Duggan, Paula S.; Gottardello, Priscila; Adams, David G. (2007). "Molecular Analysis of Genes in Nostoc punctiforme Involved in Pilus Biogenesis and Plant Infection". Journal of Bacteriology. 189 (12): 4547–4551. doi:10.1128/JB.01927-06. PMC 1913353. PMID 17416648.
- Risser, Douglas D.; Chew, William G.; Meeks, John C. (2014). "Genetic characterization of thehmplocus, a chemotaxis-like gene cluster that regulates hormogonium development and motility in Nostoc punctiforme". Molecular Microbiology. 92 (2): 222–233. doi:10.1111/mmi.12552. PMID 24533832. S2CID 37479716.
- Khayatan, Behzad; Meeks, John C.; Risser, Douglas D. (2015). "Evidence that a modified type IV pilus-like system powers gliding motility and polysaccharide secretion in filamentous cyanobacteria". Molecular Microbiology. 98 (6): 1021–1036. doi:10.1111/mmi.13205. PMID 26331359. S2CID 8749419.
- Schuergers, Nils; Nürnberg, Dennis J.; Wallner, Thomas; Mullineaux, Conrad W.; Wilde, Annegret (2015). "PilB localization correlates with the direction of twitching motility in the cyanobacterium Synechocystis sp. PCC 6803". Microbiology. 161 (5): 960–966. doi:10.1099/mic.0.000064. PMID 25721851.
- Halfen, Lawrence N.; Castenholz, Richard W. (1970). "Gliding in a Blue–Green Alga: A Possible Mechanism". Nature. 225 (5238): 1163–1165. Bibcode:1970Natur.225.1163H. doi:10.1038/2251163a0. PMID 4984867. S2CID 10399610.
- Halfen, Lawrence N.; Castenholz, Richard W. (1971). "Gliding Motility in the Blue-Green Alga Oscillatoria Princeps 1". Journal of Phycology. 7 (2): 133–145. doi:10.1111/j.1529-8817.1971.tb01492.x. S2CID 86115246.
- Tchoufag, Joël; Ghosh, Pushpita; Pogue, Connor B.; Nan, Beiyan; Mandadapu, Kranthi K. (2019). "Mechanisms for bacterial gliding motility on soft substrates". Proceedings of the National Academy of Sciences. 116 (50): 25087–25096. arXiv:1807.07529. Bibcode:2019PNAS..11625087T. doi:10.1073/pnas.1914678116. PMC 6911197. PMID 31767758.
- Shepard, R. N.; Sumner, D. Y. (2010). "Undirected motility of filamentous cyanobacteria produces reticulate mats". Geobiology. 8 (3): 179–190. Bibcode:2010Gbio....8..179S. doi:10.1111/j.1472-4669.2010.00235.x. PMID 20345889. S2CID 24452272.
- Davies, Neil S.; Liu, Alexander G.; Gibling, Martin R.; Miller, Randall F. (2016). "Resolving MISS conceptions and misconceptions: A geological approach to sedimentary surface textures generated by microbial and abiotic processes". Earth-Science Reviews. 154: 210–246. Bibcode:2016ESRv..154..210D. doi:10.1016/j.earscirev.2016.01.005. S2CID 56345018.
- Allwood, Abigail C.; Walter, Malcolm R.; Kamber, Balz S.; Marshall, Craig P.; Burch, Ian W. (2006). "Stromatolite reef from the Early Archaean era of Australia". Nature. 441 (7094): 714–718. Bibcode:2006Natur.441..714A. doi:10.1038/nature04764. PMID 16760969. S2CID 4417746.
- Sumner, Dawn Y. (1997). "Late Archean Calcite-Microbe Interactions: Two Morphologically Distinct Microbial Communities That Affected Calcite Nucleation Differently". PALAIOS. 12 (4): 302–318. Bibcode:1997Palai..12..302S. doi:10.2307/3515333. JSTOR 3515333.
- Tamulonis, Carlos; Kaandorp, Jaap (2014). "A Model of Filamentous Cyanobacteria Leading to Reticulate Pattern Formation". Life. 4 (3): 433–456. Bibcode:2014Life....4..433T. doi:10.3390/life4030433. PMC 4206854. PMID 25370380.
- Shaw, T.; Winston, M.; Rupp, C. J.; Klapper, I.; Stoodley, P. (2004). "Commonality of Elastic Relaxation Times in Biofilms". Physical Review Letters. 93 (9): 098102. Bibcode:2004PhRvL..93i8102S. doi:10.1103/PhysRevLett.93.098102. PMID 15447143.
- Tamulonis, Carlos; Postma, Marten; Kaandorp, Jaap (2011). "Modeling Filamentous Cyanobacteria Reveals the Advantages of Long and Fast Trichomes for Optimizing Light Exposure". PLOS ONE. 6 (7): e22084. Bibcode:2011PLoSO...622084T. doi:10.1371/journal.pone.0022084. PMC 3138769. PMID 21789215.
- Donkor, Victoria A.; Amewowor, Damina H.A.K.; Hã¤Der, Donat-P. (1993). "Effects of tropical solar radiation on the motility of filamentous cyanobacteria". FEMS Microbiology Ecology. 12 (2): 143–147. doi:10.1111/j.1574-6941.1993.tb00026.x.
- Garcia-Pichel, Ferran; Mechling, Margaret; Castenholz, Richard W. (1994). "Diel Migrations of Microorganisms within a Benthic, Hypersaline Mat Community". Applied and Environmental Microbiology. 60 (5): 1500–1511. Bibcode:1994ApEnM..60.1500G. doi:10.1128/aem.60.5.1500-1511.1994. PMC 201509. PMID 16349251.
- Fourã§Ans, Aude; Solã©, Antoni; Diestra, Ella; Ranchou-Peyruse, Anthony; Esteve, Isabel; Caumette, Pierre; Duran, Robert (2006). "Vertical migration of phototrophic bacterial populations in a hypersaline microbial mat from Salins-de-Giraud (Camargue, France)". FEMS Microbiology Ecology. 57 (3): 367–377. doi:10.1111/j.1574-6941.2006.00124.x. PMID 16907751.
- Richardson, Laurie L.; Castenholz, Richard W. (1987). "Diel Vertical Movements of the Cyanobacterium Oscillatoria terebriformis in a Sulfide-Rich Hot Spring Microbial Mat". Applied and Environmental Microbiology. 53 (9): 2142–2150. Bibcode:1987ApEnM..53.2142R. doi:10.1128/aem.53.9.2142-2150.1987. PMC 204072. PMID 16347435.
- McBride, Mark J. (2001). "Bacterial Gliding Motility: Multiple Mechanisms for Cell Movement over Surfaces". Annual Review of Microbiology. 55: 49–75. doi:10.1146/annurev.micro.55.1.49. PMID 11544349.
- Reichenbach, H. (1981). "Taxonomy of the Gliding Bacteria". Annual Review of Microbiology. 35: 339–364. doi:10.1146/annurev.mi.35.100181.002011. PMID 6794424.
- Hoiczyk, Egbert; Baumeister, Wolfgang (1998). "The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria". Current Biology. 8 (21): 1161–1168. doi:10.1016/S0960-9822(07)00487-3. PMID 9799733. S2CID 14384308.
- Hoiczyk, E. (2000). "Gliding motility in cyanobacteria: Observations and possible explanations". Archives of Microbiology. 174 (1–2): 11–17. doi:10.1007/s002030000187. PMID 10985737. S2CID 9927312.
- Bhaya, D.; Watanabe, N.; Ogawa, T.; Grossman, A. R. (1999). "The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. Strain PCC6803". Proceedings of the National Academy of Sciences. 96 (6): 3188–3193. Bibcode:1999PNAS...96.3188B. doi:10.1073/pnas.96.6.3188. PMC 15917. PMID 10077659.
- Hoiczyk, E.; Baumeister, W. (1995). "Envelope structure of four gliding filamentous cyanobacteria". Journal of Bacteriology. 177 (9): 2387–2395. doi:10.1128/jb.177.9.2387-2395.1995. PMC 176896. PMID 7730269.
- Halfen, Lawrence N.; Castenholz, Richard W. (1971). "Gliding Motility in the Blue-Green Alga Oscillatoria Princeps 1". Journal of Phycology. 7 (2): 133–145. doi:10.1111/j.1529-8817.1971.tb01492.x. S2CID 86115246.
- "Enhanced Model for Photophobic Responses of the Blue-Green Alga, <italic>Phormidium uncinatum</italic>". Plant and Cell Physiology. 1982. doi:10.1093/oxfordjournals.pcp.a076487.
- Häder, Donat-P. (1987). "EFFECTS OF UV-B IRRADIATION ON PHOTOMOVEMENT IN THE DESMID, Cosmarium cucumis". Photochemistry and Photobiology. 46: 121–126. doi:10.1111/j.1751-1097.1987.tb04745.x. S2CID 97100233.
- Gabai, V.L. (1985). "A one-instant mechanism of phototaxis in the cyanobacterium Phormidium uncinatum". FEMS Microbiology Letters. 30 (1–2): 125–129. doi:10.1111/j.1574-6968.1985.tb00998.x.
- Häder, D.P. (1987) "Photomovement". The Cyanobacteria: 325-345.
- Castenholz, Richard W. (2015-09-14), "Cyanobacteria", Cyanobacteria, Wiley, pp. 1–2, doi:10.1002/9781118960608.pbm00010, ISBN 9781118960608
- Hangarter, Roger P.; Gest, Howard (2004). "Pictorial Demonstrations of Photosynthesis". Photosynthesis Research. Springer Science and Business Media LLC. 80 (1–3): 421–425. doi:10.1023/b:pres.0000030426.98007.6a. ISSN 0166-8595. PMID 16328838. S2CID 9453250.