Fragmentation (mass spectrometry)
In mass spectrometry, fragmentation is the dissociation of energetically unstable molecular ions formed from passing the molecules mass spectrum. These reactions are well documented over the decades and fragmentation patterns are useful to determine the molar weight and structural information of unknown molecules.[1][2] Fragmentation that occurs in tandem mass spectrometry experiments has been a recent focus of research, because this data helps facilitate the identification of molecules.[3]
Mass spectrometry techniques
Fragmentation can occur in the ion source (in-source fragmentation)[4][5] where it has been used with electron ionization[4] to help identify molecules and, recently (2020), with electrospray ionization it has been shown to provide the same benefit in facilitating molecular identification.[5] Prior to these experiments,[5][6] electrospray ionization in-source fragmentation was generally considered an undesired effect[7] however, electrospray ionization using Enhanced In-Source Fragmentation/Annotation (EISA) has been shown to promote in-source fragmentation that creates fragment ions that are consistent with tandem mass spectrometers.[5][6] Tandem mass spectrometry-generated fragmentation is typically made in the collision zone (post-source fragmentation) of a tandem mass spectrometer. EISA and collision-induced dissociation (CID) among other physical events that impact ions are a part of gas-phase ion chemistry. A few different types of mass fragmentation are collision-induced dissociation (CID) through collision with neutral molecule, surface-induced dissociation (SID) using fast moving ions collision with a solid surface, laser induced dissociation which uses laser to induce the ion formation, electron-capture dissociation (ECD) due to capturing of low energy electrons, electron-transfer dissociation (ETD) through electron transfer between ions, negative electron-transfer dissociation (NETD), electron-detachment dissociation (EDD), photodissociation, particularly infrared multiphoton dissociation (IRMPD) using IR radiation for the bombardment and blackbody infrared radiative dissociation (BIRD) which use IR radiation instead of laser, higher-energy C-trap dissociation (HCD), EISA, and charge remote fragmentation.[8][9][10]
Fragmentation reactions
Fragmentation is a type of chemical dissociation, in which the removal of the electron from the molecule results in ionization. Removal of electrons from either sigma bond, pi bond or nonbonding orbitals causes the ionization.[2] This can take place by a process of homolytic cleavage or homolysis or heterolytic cleavage or heterolysis of the bond. Relative bond energy and the ability to undergo favorable cyclic transition states affect the fragmentation process. Rules for the basic fragmentation processes are given by Stevenson's Rule.
_V.1.svg.png.webp)
.svg.png.webp)
Two major categories of bond cleavage patterns are simple bond cleavage reactions and rearrangement reactions.[2]
Simple bond cleavage reactions
Majority of organic compounds undergo simple bond cleavage reactions, in which direct cleavage of bond take place. Sigma bond cleavage, radical site-initiated fragmentation, and charge site-initiated fragmentation are few types of simple bond cleavage reactions.[2]
Sigma bond cleavage / σ-cleavage
Sigma bond cleavage is most commonly observed in molecules, which can produce stable cations such as saturated alkanes, secondary and tertiary carbocations. This occurs when an alpha electron is removed. The C-C bond elongates and weakens causing fragmentation. Fragmentation at this site produces a charged and a radical fragment.[2]
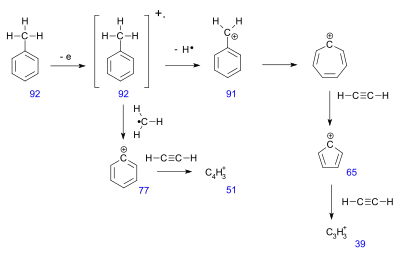
Radical site-initiated fragmentation
Sigma bond cleavage also occurs on radical cations remote from the site of ionization. This is commonly observed in alcohols, ethers, ketones, esters, amines, alkenes, and aromatic compounds with a carbon attached to ring. The cation has a radical on a heteroatom or an unsaturated functional group. The driving force of fragmentation is the strong tendency of the radical ion for electron pairing. Cleavage occurs when the radical and an odd electron from the bonds adjacent to the radical migrate to form a bond between the alpha carbon and either the heteroatom or the unsaturated functional group. The sigma bond breaks; hence this cleavage is also known as homolytic bond cleavage or α-cleavage.[2]
Charge site-initiated cleavage
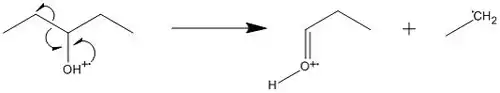
The driving force of charge site-initiated fragmentation is the inductive effect of the charge site in radical cations. The electrons from the bond adjacent to the charge-bearing atom migrate to that atom, neutralizing the original charge and causing it to move to a different site. This term is also called inductive cleavage and is an example of heterolytic bond cleavage.[2]
Rearrangement reactions
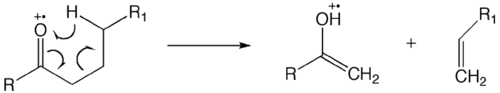
Rearrangement reactions are fragmentation reactions that form new bonds producing an intermediate structure before cleavage. One of the most studied rearrangement reaction is the McLafferty rearrangement / γ-hydrogen rearrangement. This occurs in the radical cations with unsaturated functional groups, like ketones, aldehydes, carboxylic acids, esters, amides, olefins, phenylalkanes. During this reaction, γ-hydrogen will transfer to the functional group at first and then subsequent α, β-bond cleavage of the intermediate will take place.[2] Other rearrangement reactions include heterocyclic ring fission (HRF), benzofuran forming fission (BFF), quinone methide (QM) fission or Retro Diels-Alder (RDA).[11]
See also
References
- McLafferty FW (1 January 1993). Interpretation of Mass Spectra. University Science Books. ISBN 978-0-935702-25-5.
- Dass C (2007). Fundamentals of contemporary mass spectrometry ([Online-Ausg.]. ed.). Hoboken, NJ [u.a.]: Wiley. ISBN 978-0-471-68229-5.
- Xue J, Guijas C, Benton HP, Warth B, Siuzdak G (October 2020). "2 molecular standards database: a broad chemical and biological resource". Nature Methods. 17 (10): 953–954. doi:10.1038/s41592-020-0942-5. PMC 8802982. PMID 32839599. S2CID 221285246.
- Gohlke RS, McLafferty FW (1993-05-01). "Early gas chromatography/mass spectrometry". Journal of the American Society for Mass Spectrometry. 4 (5): 367–371. doi:10.1016/1044-0305(93)85001-E. PMID 24234933. S2CID 33972992.
- Xue J, Domingo-Almenara X, Guijas C, Palermo A, Rinschen MM, Isbell J, et al. (April 2020). "Enhanced in-Source Fragmentation Annotation Enables Novel Data Independent Acquisition and Autonomous METLIN Molecular Identification". Analytical Chemistry. 92 (8): 6051–6059. doi:10.1021/acs.analchem.0c00409. PMC 8966047. PMID 32242660. S2CID 214768212.
- Domingo-Almenara X, Montenegro-Burke JR, Guijas C, Majumder EL, Benton HP, Siuzdak G (March 2019). "Autonomous METLIN-Guided In-source Fragment Annotation for Untargeted Metabolomics". Analytical Chemistry. 91 (5): 3246–3253. doi:10.1021/acs.analchem.8b03126. PMC 6637741. PMID 30681830.
- Lu W, Su X, Klein MS, Lewis IA, Fiehn O, Rabinowitz JD (June 2017). "Metabolite Measurement: Pitfalls to Avoid and Practices to Follow". Annual Review of Biochemistry. 86 (1): 277–304. doi:10.1146/annurev-biochem-061516-044952. PMC 5734093. PMID 28654323.
- Yost RA, Enke CG (1978). "Selected ion fragmentation with a tandem quadrupole mass spectrometer". Journal of the American Chemical Society. 100 (7): 2274–2275. doi:10.1021/ja00475a072.
- Lermyte F, Valkenborg D, Loo JA, Sobott F (November 2018). "Radical solutions: Principles and application of electron-based dissociation in mass spectrometry-based analysis of protein structure" (PDF). Mass Spectrometry Reviews. 37 (6): 750–771. doi:10.1002/mas.21560. PMC 6131092. PMID 29425406.
- Chen X, Wang Z, Wong YE, Wu R, Zhang F, Chan TD (November 2018). "Electron-ion reaction-based dissociation: A powerful ion activation method for the elucidation of natural product structures". Mass Spectrometry Reviews. 37 (6): 793–810. doi:10.1002/mas.21563. PMID 29603345.
- Li HJ, Deinzer ML (February 2007). "Tandem mass spectrometry for sequencing proanthocyanidins". Analytical Chemistry. 79 (4): 1739–48. doi:10.1021/ac061823v. PMID 17297981.