Ghost lineage
A ghost lineage is a hypothesized ancestor in a species lineage that has left no fossil evidence, but can still be inferred to exist or have existed because of gaps in the fossil record or genomic evidence.[1][2] The process of determining a ghost lineage relies on fossilized evidence before and after the hypothetical existence of the lineage and extrapolating relationships between organisms based on phylogenetic analysis.[3] Ghost lineages assume unseen diversity in the fossil record and serve as predictions for what the fossil record could eventually yield; these hypotheses can be tested by unearthing new fossils or running phylogenetic analyses.[4]
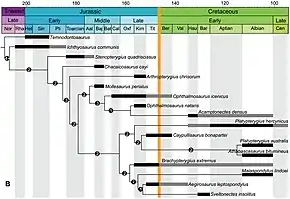
Ghost lineages and Lazarus taxa are related concepts, as both stem from gaps in the fossil record.[2] A ghost lineage is any gap in a taxon's fossil record, with or without reappearance, while a Lazarus taxon is a type of ghost lineage wherein a species is believed to have gone extinct due to an absence of it in the fossil record, but then reappears after a period of time.[2] Examples of Lazarus taxa include the famous coelacanths, as well as the Philippine naked-backed fruit bat.[5]
Name
In 1992, an article stated: "These additional entities are taxa [groups] that are predicted to occur by the internal branching structure of phylogenetic trees....I refer to these as ghost lineages because they are invisible to the fossil record."[6] Phylogenetic trees constructed based on fossil records and Darwin's Theory of Evolution often give an indication that species with similar phenotypes existed, although its fossil has not been discovered.[7]
It is important to note that ghost lineages and ghost taxa are not the same. A ghost lineage is a one direct connection between the descendant and the ancestor, whereas a ghost taxon has many split descendants.[3]
Examples

When looking back at extinct organisms, there are some groups of organisms (or lineages) that have gaps in their fossil records. These organisms or species may be closely related to one another, but there are no traces in the fossil records or sediment beds that might shed some light on their origins. Biologists may infer the existence of ghost lineages by examining sequential stratigraphic units in the fossil record.[8] Fossils can then be mapped onto cladograms and range charts to assess which lineages are missing in the fossil record.[8] A classic example is the coelacanth, a type of fish related to the lungfishes and to primitive tetrapods. It seems that coelacanths have also been around for the past 80 million years but have failed to leave any fossils. The reason for this is their environment, which is deep water near volcanic islands; therefore, these sediments are hard to get to, giving these coelacanths an 80 million year gap or ghost lineage.[2] Another ghost lineage was that of the averostran theropods, a ghost lineage now reduced considerably due to the discovery of Tachiraptor.[9]
Duration and diversification
The duration between distinct fossils can be measured and used to derive information about the evolutionary history of a species. A study conducted in 1998 showed that a correlation exists between the diversification of a species and the duration of its ghost lineage; namely that a shorter ghost lineage implies that there will be greater species diversification.[10]
Genetic evidence
Genetic evidence has revealed ghost populations in many species, including modern bonobos and chimpanzees, allopolypoid frogs, polyploid parthenogenetic crayfish, a variety of plants, and humans.[11][12][13] A study comparing the genomes of 69 modern bonobos and chimpanzees found between .9-4.2% of gene flow from ancient bonobos and an archaic great ape lineage to modern bonobos, allowing researchers to reconstruct 4.8% of this ghost population’s genome.[11] Furthermore, previous models for European ancestry suggested that European populations descended from two ancient populations, but genetic evidence now suggests that a third “ghost population”, the Ancient North Eurasians, has also contributed to European ancestry.[13]
See also
References
- Lorente-Galdos B, Lao O, Serra-Vidal G, Santpere G, Kuderna LF, Arauna LR, et al. (April 2019). "Whole-genome sequence analysis of a Pan African set of samples reveals archaic gene flow from an extinct basal population of modern humans into sub-Saharan populations". Genome Biology. 20 (1): 77. doi:10.1186/s13059-019-1684-5. PMC 6485163. PMID 31023378.
- Wedel M (May 2010). "Ghost lineages". University of California Museum of Paleontology. Retrieved November 4, 2019.
- Norell MA (January 1993). "Tree-Based Approaches to Understanding History; Comments on Ranks, Rules and the Quality of the Fossil Record" (PDF). American Journal of Science. 293 (A): 407–417. Bibcode:1993AmJS..293..407N. doi:10.2475/ajs.293.a.407.
- Dingus L, Rowe T (1998). The Mistaken Extinction: Dinosaur Evolution and the Origin of Birds. New York: W. H. Freeman.
- Gentle L. "Meet the Lazarus creatures – six species we thought were extinct, but aren't". The Conversation. Retrieved 2019-12-04.
- Norell MA (1992). Novacek MJ, Wheeler QD (eds.). Taxic Origin and Temporal Diversity: The Effect of Phylogeny, in Extinction and Phylogeny. Columbia University Press, New York. pp. 89–118.
- Norell MA, Novacek MJ (March 1992). "The fossil record and evolution: comparing cladistic and paleontologic evidence for vertebrate history". Science. 255 (5052): 1690–3. Bibcode:1992Sci...255.1690N. doi:10.1126/science.255.5052.1690. PMID 17749423. S2CID 26752987.
- Wills MA (October 2007). "Fossil ghost ranges are most common in some of the oldest and some of the youngest strata". Proceedings. Biological Sciences. 274 (1624): 2421–7. doi:10.1098/rspb.2007.0357. PMC 2274967. PMID 17652067.
- Langer MC, Rincón AD, Ramezani J, Solórzano A, Rauhut OW (October 2014). "New dinosaur (Theropoda, stem-Averostra) from the earliest Jurassic of the La Quinta formation, Venezuelan Andes". Royal Society Open Science. Royal Society. 1 (2): 140184. Bibcode:2014RSOS....140184L. doi:10.1098/rsos.140184. PMC 4448901. PMID 26064540.
- Sidor CA, Hopson JA (1998). "Ghost lineages and "mammalness": assessing the temporal pattern of character acquisition in the Synapsida". Paleobiology. 24 (2): 254–273. doi:10.1666/0094-8373(1998)024[0254:GLAATT]2.3.CO;2. S2CID 83773704.
- Kuhlwilm M, Han S, Sousa VC, Excoffier L, Marques-Bonet T (June 2019). "Ancient admixture from an extinct ape lineage into bonobos". Nature Ecology & Evolution. 3 (6): 957–965. doi:10.1038/s41559-019-0881-7. PMID 31036897. S2CID 139107044.
- Unmack PJ, Adams M, Bylemans J, Hardy CM, Hammer MP, Georges A (March 2019). "Perspectives on the clonal persistence of presumed 'ghost' genomes in unisexual or allopolyploid taxa arising via hybridization". Scientific Reports. 9 (1): 4730. Bibcode:2019NatSR...9.4730U. doi:10.1038/s41598-019-40865-3. PMC 6426837. PMID 30894575.
- "New Branch Added to European Family Tree". Harvard Medical School. Retrieved 2019-12-11.