LRRC57
Leucine rich repeat containing 57, also known as LRRC57 is a protein encoded in humans by the LRRC57 gene.[5]
| LRRC57 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | LRRC57, leucine rich repeat containing 57 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1913856 HomoloGene: 11995 GeneCards: LRRC57 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Function
The exact function of LRRC57 is not known. It is a member of the leucine-rich repeat family of proteins, which are known to be involved in protein-protein interactions.
Protein sequence
As is customary for leucine-rich repeat proteins,[6] the sequence[5] is shown below with the repeats starting on their own lines. The beginning of each repeat is a β-strand, which forms a β-sheet along the concave side of the protein. The convex side of the protein is formed by the latter half of each repeat, and may consist of a variety of structures, including α-helices, 310 helices, β-turns, and even short β-strands.[6]
Note that the 5' and 3' UTR both are rich in leucines, suggesting that they may be degenerate repeats (the overall protein is 19.7% leucine and 7.5% asparagine, both very rich).
The following layout of the LRRC57 amino acid sequence makes it easy to discern the LxxLxLxxNxxL consensus sequence of LRRs.[6]
1 M G N S A '''<span style="color:orange">L</span>''' R A H V E T A Q K T G V F Q '''<span style="color:orange">L</span>''' K D R G L T E F P A D L Q K L T S N 39
40 '''<span style="color:orange">L</span>''' R T I D '''<span style="color:orange">L</span>''' S N '''<span style="color:orange">N</span>''' K I E S '''<span style="color:orange">L</span>''' P P L L I G K F T L 63
64 '''<span style="color:orange">L</span>''' K S '''<span style="color:orange">L</span>''' S '''<span style="color:orange">L</span>''' N N '''<span style="color:orange">N</span>''' K '''<span style="color:orange">L</span>''' T V '''<span style="color:orange">L</span>''' P D E I C N '''<span style="color:orange">L</span>''' K K 86
87 '''<span style="color:orange">L</span>''' E T <span style="color:orange">L</span> S <span style="color:orange">L</span> N N <span style="color:blue">N</span> H <span style="color:orange">L</span> R E <span style="color:orange">L</span> P S T F G Q <span style="color:orange">L</span> S A 109
110 <span style="color:orange">L</span> K T <span style="color:orange">L</span> S <span style="color:orange">L</span> S G <span style="color:blue">N</span> Q <span style="color:orange">L</span> G A <span style="color:orange">L</span> P P Q L C S <span style="color:orange">L</span> R H 132
133 <span style="color:orange">L</span> D V M D <span style="color:orange">L</span> S K <span style="color:blue">N</span> Q I R S I P D S V G E <span style="color:orange">L</span> Q 154
155 V I E <span style="color:orange">L</span> N <span style="color:orange">L</span> N Q <span style="color:blue">N</span> Q I S Q I S V K I S C C P R 177
178 <span style="color:orange">L</span> K I <span style="color:orange">L</span> R <span style="color:orange">L</span> E E <span style="color:blue">N</span> C <span style="color:orange">L</span> E L S M L P Q S I <span style="color:orange">L</span> S D 200
201 S Q I C L <span style="color:orange">L</span> A V E G N L F E I K K L R E <span style="color:orange">L</span> E G Y D K Y M E R F T A T K K K F A 239
<span style="color:orange">L</span> x x <span style="color:orange">L</span> x <span style="color:orange">L</span> x x <span style="color:blue">N</span> x <span style="color:orange">L</span> x x <span style="color:orange">L</span> x x x x x x <span style="color:orange">L</span> x
Homology
LRRC57 is exceedingly well conserved, as shown by the following multiple sequence alignment, prepared using ClustalX2.[7] The cyan and yellow highlights call out regions of high conservation and the repeats.
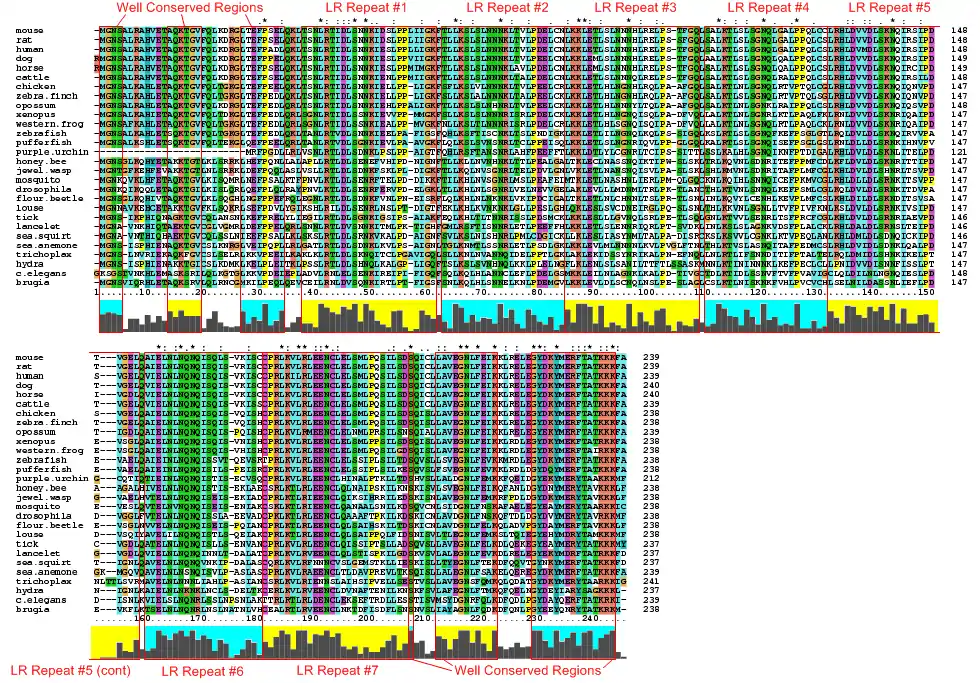
The following table provides a few details on orthologs of the human version of LRRC57. To save space, not all of these orthologs are included in the above multiple sequence alignment. These orthologs were gathered from BLAT.[8] and BLAST searches[9]
| Species | Organism common name | NCBI accession | Sequence identity | Sequence similarity | Length (AAs) | Gene common name |
| Homo sapiens | Human | NP_694992 | 100% | 100% | 239 | leucine rich repeat containing 57 |
| Pan troglodytes | Chimpanzee | XP_510338 | 99% | 100% | 165 | PREDICTED: hypothetical protein |
| Orangutan | 99% | 99% | 238 | From BLAT – no GenBank record | ||
| Macaca mulatta | Rhesus macaque | XP_001100633 | 96% | 99% | 143 | PREDICTED: similar to CG3040-PA |
| Mus musculus | House mouse | NP_079933 | 95% | 99% | 239 | leucine rich repeat containing 57 |
| Rattus norvegicus | Norway rat | NP_001012354 | 95% | 99% | 239 | leucine rich repeat containing 57 |
| Canis lupus familiaris | Dog | XP_535443 | 94% | 98% | 264 | PREDICTED: similar to CG3040-PA |
| Equus caballus | Horse | XP_001503298 | 94% | 97% | 273 | PREDICTED: similar to leucine rich repeat containing 57 |
| Bos taurus | Cattle | NP_001026924 | 94% | 97% | 239 | leucine rich repeat containing 57 |
| Monodelphis domestica | Opossum | XP_001362682 | 84% | 94% | 239 | PREDICTED: hypothetical protein |
| Ornithorhynchus anatinus | Platypus | XP_001520403 | 76% | 92% | 99 | PREDICTED: hypothetical protein |
| Gallus gallus | Chicken | XP_421160 | 85% | 92% | 238 | PREDICTED: hypothetical protein |
| Taeniopygia guttata | Zebra finch | XP_002200369 | 85% | 92% | 238 | PREDICTED: leucine rich repeat containing 57 |
| Xenopus laevis | African clawed frog | NP_001085208 | 76% | 88% | 238 | hypothetical protein LOC432302 |
| Xenopus (Silurana) tropicalis | Western clawed frog | NP_001120199 | 76% | 87% | 238 | hypothetical protein LOC100145243 |
| Danio rerio | Zebrafish | NP_001002627 | 69% | 83% | 238 | leucine rich repeat containing 57 |
| Tetraodon nigroviridis | Spotted green pufferfish | CAF89640 | 67% | 83% | 238 | unnamed protein product |
| Branchiostoma floridae | Florida lancelet | XP_002209325 | 57% | 78% | 237 | hypothetical protein BRAFLDRAFT_277364 |
| Ciona intestinalis | (a sea squirt) | XP_002129992 | 50% | 71% | 237 | PREDICTED: similar to Leucine rich repeat containing 57 |
| Strongylocentrotus purpuratus | Purple urchin | XP_782986 | 57% | 74% | 212 | PREDICTED: hypothetical protein |
| Ixodes scapularis | Black-legged tick | EEC17869 | 57% | 73% | 237 | leucine rich domain-containing protein, putative |
| Apis mellifera | Honey bee | XP_001121818 | 53% | 72% | 238 | PREDICTED: similar to CG3040-PA |
| Nasonia vitripennis | Jewel wasp | XP_001601190 | 57% | 73% | 238 | PREDICTED: similar to ENSANGP00000011808 |
| Tribolium castaneum | Red flour beetle | XP_973486 | 56% | 70% | 238 | PREDICTED: similar to AGAP001491-PA |
| Pediculus humanus | Body louse | EEB17844 | 52% | 72% | 238 | leucine-rich repeat-containing protein, putative |
| Aedes aegypti | Yellow fever mosquito | XP_001657420 | 50% | 66% | 239 | internalin A |
| Culex quinquefasciatus | Southern house mosquito | XP_001865691 | 49% | 67% | 238 | leucine-rich repeat-containing protein 57 |
| Drosophila melanogaster | Fruit fly | NP_572372 | 50% | 67% | 238 | CG3040 |
| Drosophila simulans | XP_002106344 | 49% | 67% | 238 | GD16172 | |
| Drosophila sechellia | XP_002043192 | 49% | 67% | 238 | GM17488 | |
| Drosophila yakuba | XP_002101312 | 50% | 68% | 238 | GE17554 | |
| Drosophila erecta | XP_001978503 | 50% | 67% | 238 | GG17646 | |
| Drosophila ananassae | XP_001964158 | 51% | 68% | 238 | GF20868 | |
| Drosophila pseudoobscura | XP_001355271 | 49% | 66% | 238 | GA15818 | |
| Drosophila persimilis | XP_002025298 | 49% | 66% | 238 | GL13411 | |
| Drosophila virilis | XP_002056963 | 51% | 68% | 238 | GJ16607 | |
| Drosophila mojavensis | XP_002010408 | 51% | 68% | 238 | GI14698 | |
| Drosophila grimshawi | XP_001991745 | 52% | 68% | 238 | GH12826 | |
| Drosophila willistoni | XP_002071645 | 50% | 67% | 238 | GK10093 | |
| Anopheles gambiae | XP_321630 | 46% | 66% | 238 | AGAP001491-PA | |
| Caenorhabditis elegans | (a nematode) | NP_740983 | 43% | 63% | 485 | hypothetical protein ZK546.2 |
| Caenorhabditis briggsae | (a nematode) | XP_001679881 | 41% | 64% | 439 | Hypothetical protein CBG02285 |
Gene neighborhood
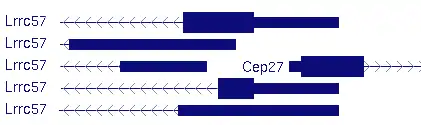
The LRRC57 gene has interesting relationships to its neighbors – HAUS2 upstream and SNAP23 downstream, as shown below for human.[10]

Shown below is the neighborhood for the mouse[11] ortholog. Note that the neighbors are the same, which is true for most vertebrates.

Note the close proximity between LRRC57 and HAUS2/CEP27 (the same gene by different names). In humans, the exons are 50bp apart, whereas in mouse, they overlap, as shown in the closeup, below. This close relationship may partially explain the high conservation of LRRC57, as it would require a mutation to be stable in both genes at the same time.
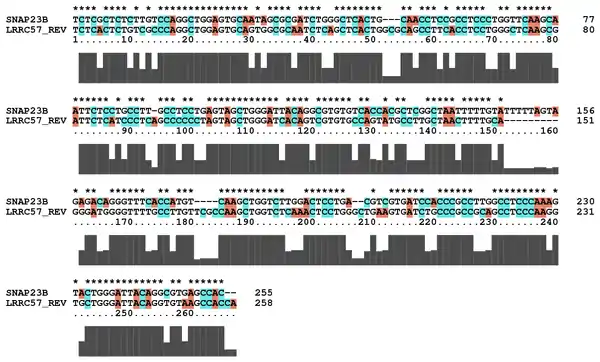
The relationship to the downstream neighbor, SNAP23 is also interesting. Quoting from the AceView[12] entry: "373 bp of this gene are antisense to spliced gene SNAP23, raising the possibility of regulated alternate expression". Taking the reverse complement of the LRRC57 cDNA and aligning it with the SNAP23 cDNA does show high similarity, as shown in this partial alignment:
Predicted post-translational modifications
The tools on the ExPASy Proteomics site[13] predict the following post-translational modifications:
| Tool | Predicted Modification | Homo sapiens | Mus musculus | Gallus gallus | Drosophila melanogaster |
| YinOYang[14] | O-β-GlcNAc | S166 | S166 | S165 | T16, T102 |
| NetPhos[15] | phosphorylation | S145, S149, S169, S199, S201, T27 T234 | S139, S145, S169, S199, S201, T27, T149, T234 | S148, S198, S200, T22 | S46, S69, S200, T179, T193, Y230 |
| Sulfinator[16] | sulfation | Y224, Y227 | Y224, Y227 | Y223, Y226 | (none) |
| SulfoSite[17] | sulfation | Y224 | Y224 | Y223 | Y223 |
| SumoPlot[18] | sumoylation | K86, K15, and K236 | (not checked) | (not checked) | (not checked) |
| Terminator[19] | N-terminus | G2 | G2 | G2 | G2 |
The predicted modifications for Homo sapiens are shown on the following conceptual translation. The cyan highlights are predicted phosphorylation sites and the yellow highlights are as labeled. The red boxes show predictions that are conserved across all four organisms.

The sites for all four organisms are highlighted on the following multiple sequence alignment.
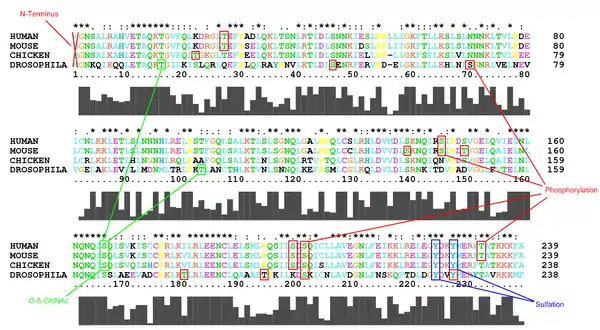
Note that the phosphorylation at S201 and the sulfation at Y224 are the only well conserved predictions across all four organisms.
Structure
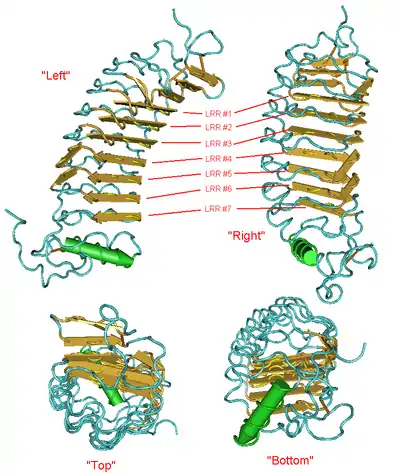
The structure of LRRC57 is not known. However, a protein BLAST search against the protein databank returns a similar protein (PDB: 2O6Q), with an E-value of 3E−14. It is also a leucine rich repeat containing seven repeats of the same length as LRRC57, described as Eptatretus burgeri (inshore hagfish) variable lymphocyte receptors A29.[22]
References
- GRCh38: Ensembl release 89: ENSG00000180979 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000027286 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: LRRC57 leucine rich repeat containing 57". Retrieved 4 May 2009.
- Bella J, Hindle KL, McEwan PA, Lovell SC (August 2008). "The leucine-rich repeat structure". Cellular and Molecular Life Sciences. 65 (15): 2307–33. doi:10.1007/s00018-008-8019-0. PMID 18408889. S2CID 10222798.
- "Clustal Home Page". Retrieved 4 May 2009.
- "BLAT Search Genome". Retrieved 4 May 2009.
- "BLAST". Retrieved 4 May 2009.
- "Human (Homo sapiens) Genome Browser Gateway". Retrieved 27 Apr 2009.
- "Mouse (Mus musculus) Genome Browser Gateway". Retrieved 27 Apr 2009.
- "AceView: Homo sapiens gene LRRC57, encoding leucine rich repeat containing 57". Retrieved 1 May 2009.
- "ExPASy Proteomics tools". Retrieved 24 Apr 2009.
- "YinOYang". Retrieved 24 Apr 2009.
- "NetPhos". Retrieved 24 Apr 2009.
- "Sulfinator". Retrieved 24 Apr 2009.
- "SulfoSite". Archived from the original on 24 July 2008. Retrieved 24 Apr 2009.
- "SumoPlot". Archived from the original on 20 April 2009. Retrieved 24 Apr 2009.
- "Terminator". Archived from the original on 16 April 2008. Retrieved 24 Apr 2009.
- "Cn3D Home Page". Cn3D. National Center for Biotechnology Information, United States National Institutes of Health. 2008-04-24. Retrieved 2009-05-06.
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH (June 2000). "Cn3D: sequence and structure views for Entrez". Trends in Biochemical Sciences. 25 (6): 300–2. doi:10.1016/S0968-0004(00)01561-9. PMID 10838572.
- Kim HM, Oh SC, Lim KJ, Kasamatsu J, Heo JY, Park BS, Lee H, Yoo OJ, Kasahara M, Lee JO (2007). "Structural diversity of the hagfish variable lymphocyte receptors". J Biol Chem. 282 (9): 6726–32. doi:10.1074/jbc.M608471200. PMID 17192264.



