Microsomal epoxide hydrolase
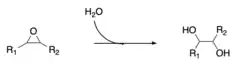
In enzymology, a microsomal epoxide hydrolase (mEH) (EC 3.3.2.9) is an enzyme that catalyzes the hydrolysis reaction between an epoxide and water to form a diol.
| Microsomal epoxide hydrolase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 A cartoon depiction of microsomal epoxide hydrolase from Aspergillus niger | |||||||||
| Identifiers | |||||||||
| EC no. | 3.3.2.9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
This enzyme plays a role in the uptake of bile salts within the large intestine. It functions as a Na+ dependent transporter. This enzyme participates in metabolism of xenobiotics by cytochrome p450. mEH has been identified as playing a large role in the detoxification and bioactivation of a wide variety of substrates, such as polycyclic aromatic hydrocarbons (PAHs), which are known for their carcinogenic properties.[1]
The human homolog of microsomal epoxide hydrolase is EPHX1 and is located on chromosome 1.[2]
Nomenclature
This enzyme belongs to the family of hydrolases, specifically those acting on ether bonds (ether hydrolases). The systematic name of this enzyme class is cis-stilbene-oxide hydrolase. Other names in common use include epoxide hydratase (ambiguous), microsomal epoxide hydratase (ambiguous), epoxide hydrase, microsomal epoxide hydrase, arene-oxide hydratase (ambiguous), benzo[a]pyrene-4,5-oxide hydratase, benzo(a)pyrene-4,5-epoxide hydratase, aryl epoxide hydrase (ambiguous), cis-epoxide hydrolase, and mEH.
Structure
Microsomal epoxide hydrolase is a single polypeptide chain composed of 455 amino acids with a molecular weight of 52.96 kilodaltons. It is known that the N-terminal region of the enzyme is responsible for anchoring the protein to the cell membrane,[3] while the C-terminal region of the enzyme contains catalytic residues.[4] Microsomal epoxide hydrolase belongs to the superfamily α/β-hydrolase fold enzymes.[5] The center of all α/β-hydrolase fold enzymes is an alpha/beta-sheet consisted of 8 beta strands connected by 6 alpha helices.[6][7] The three dimensional structure of mEH has been elucidated from Aspergillus niger.[4] Although no 3D modeling has been solved for the mammalian mEH enzyme (EPHX1), the overall homology between fungal and mammalian mEH is relatively high.[8][9][10] This high homology has allowed for the elucidation overall general structure and subsequent catalytic mechanism of EPHX1 in humans by comparisons to existing structures of fungal mEH.
Mechanism
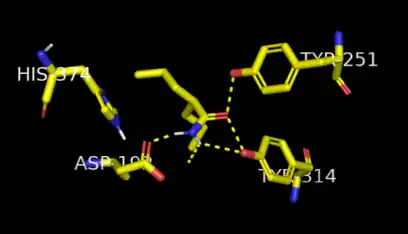
α/β-hydrolase fold enzymes use a catalytic triad in their active site. The catalytic triad present in microsomal epoxide hydrolase is composed of glutamine, histidine and aspartic acid.[10] The substrate is positioned in an orientation poised for nucleophilic attack through hydrogen bonding stabilization from two nearby tyrosine residues [11][12] The proposed mechanism for the mEH-catalyzed reaction first involves a nucleophilic attack on the oxirane ring of the substrate from the aspartic acid residue near the active site, which forms an ester intermediate.[13] The second step in this mechanism is hydrolysis of the ester that occurs by an activated water molecule.[14] The activation of water is facilitated by proton abstraction via the catalytic triad between a water molecule, glutamine, and histidine.[15] After hydrolysis, the substrate is then released from its bond to the aspartic acid residue, liberating the diol product from the enzyme active site.[16]
The active site of this enzyme lies within a hydrophobic pocket in the enzyme, which in turn leads to the enzyme's preferential reactivity with molecules with hydrophobic side-chains.[17][11] The mEH enzyme typically binds to small organic epoxides, such as styrene epoxide and cis-stillbene-oxide. mEH does not catalyze the hydrolysis of bulkier molecules, as their large side-chains may sterically disrupt the charge relay system responsible for water activation.[11]
Function
In humans, mEH has been found in the ovary, lung, kidney, lymphocytes, epithelial cells, and liver.[18] Microsomal epoxide hydrolase serves as a protective enzyme against potentially harmful small molecules derived from the external environment.[19] This hydrolysis of genotoxic epoxides causes subsequent effects in several signal transduction pathways, rendering this enzyme important to metabolism.[20][21]
Disease relevance
Microsomal epoxide hydrolase plays a large role in its effects on human health. Studies have shown that mutations EPHX1 in humans may be the cause of hypercholanemia,[22] preeclampsia,[23][24] and may contribute to fetal hydantoin syndrome.[25] Research also suggests that maternal polymorphisms in EPHX1 in pregnant women were related to facial malformations of children born from women taking phenytoin during their first trimester of pregnancy.[26] While mEH participates in the protection of human health via detoxification of various environmental substances, it also has been found to facilitate the activation of carcinogens.[1]
mEH detoxifies reactive epoxides that are commonly caused from cigarette smoke, and as such it is hypothesized that mutations in EPHX1 in humans may have an effect on an individual's susceptibility to COPD, emphysema and lung cancer. Some sources have demonstrated that individuals affected by COPD have a higher rate of containing an under-active variant of the EPHX1 gene, yet also demonstrated that the overactive variant of the gene was also found in higher frequencies in individuals affected by disease as well.[27][28] Other research has provided evidence supporting the idea that EPHX1 variants do not contribute to susceptibility of disease, but do contribute to disease severity.[1] The role that mEH plays in lung cancer and COPD is still not fully elucidated, as the data on the topic in the literature is not completely unanimous.[29]
There is some evidence that mEH variants may contribute to the occurrence of childhood asthma in combination with variants on the GSTP1 gene.[30]
Compared to soluble epoxide hydrolase, the contribution of mEH to metabolism of beneficial epoxy fatty acids such as Epoxyeicosatrienoic acid is considered minor since they are relatively poor mEH substrates in vitro. Yet, in vivo, it was found that mEH can play a considerable role in regulation of EET levels[31][32] and hence inhibition of mEH or dual inhibition of mEH and sEH might have therapeutic potential. Amide, amine and urea based mEH inhibitors have been explored.[33] Based on the most potent inhibitors characterized, an amide with a bulky alpha-substituent and a phenyl ring with lipophilic groups at meta-positions appear to be key pharmacophore units.[34]
The overall effect that mEH has on human health is still debated, with some sources finding evidence that the overactive EPHX1 gene is the culprit for some diseases, while other evidence supports that the under active variant is the cause of others.
References
- Kiyohara C, Yoshimasu K, Takayama K, Nakanishi Y (January 2006). "EPHX1 polymorphisms and the risk of lung cancer: a HuGE review". Epidemiology. 17 (1): 89–99. doi:10.1097/01.ede.0000187627.70026.23. PMID 16357600.
- Jackson MR, Craft JA, Burchell B (September 1987). "Nucleotide and deduced amino acid sequence of human liver microsomal epoxide hydrolase". Nucleic Acids Research. 15 (17): 7188. doi:10.1093/nar/15.17.7188. PMC 306212. PMID 3502697.
- Craft JA, Baird S, Lamont M, Burchell B (August 1990). "Membrane topology of epoxide hydrolase". Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism. 1046 (1): 32–9. doi:10.1016/0005-2760(90)90091-B. PMID 2397243.
- PDB: 3G0I; Zou J, Hallberg BM, Bergfors T, Oesch F, Arand M, Mowbray SL, Jones TA (February 2000). "Structure of Aspergillus niger epoxide hydrolase at 1.8 A resolution: implications for the structure and function of the mammalian microsomal class of epoxide hydrolases". Structure. 8 (2): 111–22. doi:10.1016/S0969-2126(00)00087-3. PMID 10673439.
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL (April 1992). "The α/β hydrolase fold. Protein Engineering, Design and Selection" (PDF). Protein Eng. 5 (3): 197–211. doi:10.1093/protein/5.3.197. hdl:11370/2d4c057d-1a67-437d-ad10-701f7a60f1e6. PMID 1409539.
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J (April 1992). "The alpha/beta hydrolase fold" (PDF). Protein Engineering. 5 (3): 197–211. doi:10.1093/protein/5.3.197. hdl:11370/2d4c057d-1a67-437d-ad10-701f7a60f1e6. PMID 1409539.
- Carr PD, Ollis DL (2009). "Alpha/beta hydrolase fold: an update". Protein and Peptide Letters. 16 (10): 1137–48. doi:10.2174/092986609789071298. PMID 19508187.
- Arand M, Oesch F (2002-02-14). Mammalian Xenobiotic Epoxide Hydrolases. pp. 459–483. doi:10.1002/0470846305.ch12. ISBN 9780470846308.
{{cite book}}:|work=ignored (help) - Arand M, Hemmer H, Dürk H, Baratti J, Archelas A, Furstoss R, Oesch F (November 1999). "Cloning and molecular characterization of a soluble epoxide hydrolase from Aspergillus niger that is related to mammalian microsomal epoxide hydrolase". The Biochemical Journal. 344 (1): 273–80. doi:10.1042/0264-6021:3440273. PMC 1220641. PMID 10548561.
- Arand M, Müller F, Mecky A, Hinz W, Urban P, Pompon D, Kellner R, Oesch F (January 1999). "Catalytic triad of microsomal epoxide hydrolase: replacement of Glu404 with Asp leads to a strongly increased turnover rate". The Biochemical Journal. 337 (1): 37–43. doi:10.1042/0264-6021:3370037. PMC 1219933. PMID 9854022.
- Lewis DF, Lake BG, Bird MG (June 2005). "Molecular modelling of human microsomal epoxide hydrolase (EH) by homology with a fungal (Aspergillus niger) EH crystal structure of 1.8 A resolution: structure-activity relationships in epoxides inhibiting EH activity". Toxicology in Vitro. 19 (4): 517–22. doi:10.1016/j.tiv.2004.07.001. PMID 15826809.
- Saenz-Méndez P, Katz A, Pérez-Kempner ML, Ventura ON, Vázquez M (April 2017). "Structural insights into human microsomal epoxide hydrolase by combined homology modeling, molecular dynamics simulations, and molecular docking calculations". Proteins. 85 (4): 720–730. doi:10.1002/prot.25251. PMID 28120429.
- Lacourciere GM, Armstrong RN (November 1993). "The catalytic mechanism of microsomal epoxide hydrolase involves an ester intermediate". Journal of the American Chemical Society. 115 (22): 10466–10467. doi:10.1021/ja00075a115.
- McCall PM, Srivastava S, Perry SL, Kovar DR, Gardel ML, Tirrell MV (April 2018). "Partitioning and Enhanced Self-Assembly of Actin in Polypeptide Coacervates". Biophysical Journal. 114 (7): 1636–1645. Bibcode:2018BpJ...114.1636M. doi:10.1016/j.bpj.2018.02.020. PMC 5954293. PMID 29642033.
- Oesch F, Herrero ME, Hengstler JG, Lohmann M, Arand M (May 2000). "Metabolic detoxification: implications for thresholds". Toxicologic Pathology. 28 (3): 382–7. doi:10.1177/019262330002800305. PMID 10862554.
- Reetz MT, Bocola M, Wang LW, Sanchis J, Cronin A, Arand M, Zou J, Archelas A, Bottalla AL, Naworyta A, Mowbray SL (June 2009). "Directed evolution of an enantioselective epoxide hydrolase: uncovering the source of enantioselectivity at each evolutionary stage". Journal of the American Chemical Society. 131 (21): 7334–43. doi:10.1021/ja809673d. PMID 19469578.
- Václavíková R, Hughes DJ, Souček P (October 2015). "Microsomal epoxide hydrolase 1 (EPHX1): Gene, structure, function, and role in human disease". Gene. 571 (1): 1–8. doi:10.1016/j.gene.2015.07.071. PMC 4544754. PMID 26216302.
- Bachmann K (2009). "Chapter 8: Drug Metabolism". Pharmacology. Elsevier. pp. 131–173. doi:10.1016/b978-0-12-369521-5.00008-7. ISBN 978-0-12-369521-5.
- Oesch F (May 1973). "Mammalian epoxide hydrases: inducible enzymes catalysing the inactivation of carcinogenic and cytotoxic metabolites derived from aromatic and olefinic compounds". Xenobiotica; the Fate of Foreign Compounds in Biological Systems. 3 (5): 305–40. doi:10.3109/00498257309151525. PMID 4584115.
- Samuelsson B, Dahlén SE, Lindgren JA, Rouzer CA, Serhan CN (September 1987). "Leukotrienes and lipoxins: structures, biosynthesis, and biological effects". Science. 237 (4819): 1171–6. Bibcode:1987Sci...237.1171S. doi:10.1126/science.2820055. PMID 2820055.
- Moghaddam MF, Grant DF, Cheek JM, Greene JF, Williamson KC, Hammock BD (May 1997). "Bioactivation of leukotoxins to their toxic diols by epoxide hydrolase". Nature Medicine. 3 (5): 562–6. doi:10.1038/nm0597-562. PMC 7095900. PMID 9142128.
- Zhu QS, Xing W, Qian B, von Dippe P, Shneider BL, Fox VL, Levy D (July 2003). "Inhibition of human m-epoxide hydrolase gene expression in a case of hypercholanemia". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1638 (3): 208–16. doi:10.1016/s0925-4439(03)00085-1. PMID 12878321.
- Zusterzeel PL, Rütten H, Roelofs HM, Peters WH, Steegers EA (February 2001). "Protein carbonyls in decidua and placenta of pre-eclamptic women as markers for oxidative stress". Placenta. 22 (2–3): 213–9. doi:10.1053/plac.2000.0606. PMID 11170826.
- Laasanen J, Romppanen EL, Hiltunen M, Helisalmi S, Mannermaa A, Punnonen K, Heinonen S (September 2002). "Two exonic single nucleotide polymorphisms in the microsomal epoxide hydrolase gene are jointly associated with preeclampsia". European Journal of Human Genetics. 10 (9): 569–73. doi:10.1038/sj.ejhg.5200849. PMID 12173035.
- Buehler BA, Delimont D, van Waes M, Finnell RH (May 1990). "Prenatal prediction of risk of the fetal hydantoin syndrome". The New England Journal of Medicine. 322 (22): 1567–72. doi:10.1056/NEJM199005313222204. PMID 2336087.
- Azzato EM, Chen RA, Wacholder S, Chanock SJ, Klebanoff MA, Caporaso NE (January 2010). "Maternal EPHX1 polymorphisms and risk of phenytoin-induced congenital malformations". Pharmacogenetics and Genomics. 20 (1): 58–63. doi:10.1097/fpc.0b013e328334b6a3. PMID 19952982.
- Smith CA, Harrison DJ (August 1997). "Association between polymorphism in gene for microsomal epoxide hydrolase and susceptibility to emphysema". Lancet. 350 (9078): 630–3. doi:10.1016/s0140-6736(96)08061-0. PMID 9288046.
- Brøgger J, Steen VM, Eiken HG, Gulsvik A, Bakke P (April 2006). "Genetic association between COPD and polymorphisms in TNF, ADRB2 and EPHX1". The European Respiratory Journal. 27 (4): 682–8. doi:10.1183/09031936.06.00057005. PMID 16585076.
- Postma DS, Silverman EK (2009). "Chapter 4 - Genetics of Asthma and COPD". Genetics of Asthma and COPD. Elsevier. pp. 37–51. doi:10.1016/b978-0-12-374001-4.00004-3. ISBN 9780123740014.
- Salam MT, Lin PC, Avol EL, Gauderman WJ, Gilliland FD (December 2007). "Microsomal epoxide hydrolase, glutathione S-transferase P1, traffic and childhood asthma". Thorax. 62 (12): 1050–7. doi:10.1136/thx.2007.080127. PMC 2094290. PMID 17711870.
- Marowsky A, Burgener J, Falck JR, Fritschy JM, Arand M (June 2009). "Distribution of Soluble and Microsomal Epoxide Hydrolase in the Mouse Brain and Its Contribution to Cerebral Epoxyeicosatrienoic Acid Metabolism". Neuroscience. 163 (2): 646–661. doi:10.1016/j.neuroscience.2009.06.033. PMID 19540314.
- Edin ML, Hamedani BG, Gruzdev A, Graves JP, Lih FB, Arbes SJ, Singh R, Leon AO, Bradbury JA, DeGraff LM, Hoopes SL, Arand M, Zeldin DC (January 2018). "Epoxide hydrolase 1 (EPHX1) hydrolyzes epoxyeicosanoids and impairs cardiac recovery after ischemia". The Journal of Biological Chemistry. 293 (9): 3281–3292. doi:10.1074/jbc.RA117.000298. PMC 5836130. PMID 29298899.
- Morisseau C, Newman JW, Dowdy DL, Goodrow MH, Hammock BD (April 2001). "Inhibition of Microsomal Epoxide Hydrolases by Ureas, Amides, and Amines". Chemical Research in Toxicology. 14 (4): 409–415. doi:10.1021/tx0001732. PMID 11304129.
- Barnych B, Singh N, Negrel S, Zhang Y, Magis D, Roux C, Hua X, Ding Z, Morisseau C, Tantillo DJ, Siegel JB, Hammock BD (March 2020). "Development of potent inhibitors of the human microsomal epoxide hydrolase". European Journal of Medicinal Chemistry. 193: 112206. doi:10.1016/j.ejmech.2020.112206. PMC 7366823. PMID 32203787.
Further reading
- Boyer PD, ed. (1972). The Enzymes. Vol. 7 (3rd ed.). New York: Academic Press. pp. 199–212.
- Lu AY, Ryan D, Jerina DM, Daly JW, Levin W (October 1975). "Liver microsomal expoxide hydrase. Solubilization, purification, and characterization". The Journal of Biological Chemistry. 250 (20): 8283–8. PMID 240858.
- Oesch F (April 1974). "Purification and specificity of a human microsomal epoxide hydratase". The Biochemical Journal. 139 (1): 77–88. doi:10.1042/bj1390077. PMC 1166253. PMID 4463951.
- Oesch F, Daly J (March 1971). "Solubilization, purification, and properties of a hepatic epoxide hydrase". Biochimica et Biophysica Acta (BBA) - Enzymology. 227 (3): 692–7. doi:10.1016/0005-2744(71)90018-0. PMID 4998715.
- Bellucci G, Chiappe C, Ingrosso G (1994). "Kinetics and stereochemistry of the microsomal epoxide hydrolase-catalyzed hydrolysis of cis-stilbene oxides". Chirality. 6 (7): 577–82. doi:10.1002/chir.530060711. PMID 7986671.
- Morisseau C, Hammock BD (2005). "Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles". Annual Review of Pharmacology and Toxicology. 45: 311–33. doi:10.1146/annurev.pharmtox.45.120403.095920. PMID 15822179. S2CID 28684338.
- Fretland AJ, Omiecinski CJ (December 2000). "Epoxide hydrolases: biochemistry and molecular biology". Chemico-Biological Interactions. 129 (1–2): 41–59. CiteSeerX 10.1.1.462.3157. doi:10.1016/S0009-2797(00)00197-6. PMID 11154734.
- Oesch F (May 1973). "Mammalian epoxide hydrases: inducible enzymes catalysing the inactivation of carcinogenic and cytotoxic metabolites derived from aromatic and olefinic compounds". Xenobiotica; the Fate of Foreign Compounds in Biological Systems. 3 (5): 305–40. doi:10.3109/00498257309151525. PMID 4584115.
- Lacourciere GM, Armstrong RN (1994). "Microsomal and soluble epoxide hydrolases are members of the same family of C-X bond hydrolase enzymes". Chemical Research in Toxicology. 7 (2): 121–4. doi:10.1021/tx00038a001. PMID 8199297.
- Newman JW, Morisseau C, Hammock BD (January 2005). "Epoxide hydrolases: their roles and interactions with lipid metabolism". Progress in Lipid Research. 44 (1): 1–51. doi:10.1016/j.plipres.2004.10.001. PMID 15748653.