Mitochondrial antiviral-signaling protein
Mitochondrial antiviral-signaling protein (MAVS) is a protein that is essential for antiviral innate immunity. MAVS is located in the outer membrane of the mitochondria, peroxisomes, and mitochondrial-associated endoplasmic reticulum membrane (MAM).[5][6] Upon viral infection, a group of cytosolic proteins will detect the presence of the virus and bind to MAVS, thereby activating MAVS. The activation of MAVS leads the virally infected cell to secrete cytokines. This induces an immune response which kills the host's virally infected cells, resulting in clearance of the virus.
Structure
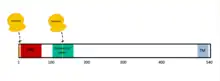
MAVS is also known as IFN-β promoter stimulator I (IPS-1), caspase activation recruitment domain adaptor inducing IFN-β(CARDIF), or virus induced signaling adaptor (VISA).[7] MAVS is encoded by a MAVS gene.[7][8] MAVS is a 540 amino acid protein that consists of three components, a N terminal caspase activation recruitment domain (CARD), a proline rich domain, and a transmembrane C terminal domain (TM).[7]
After the MAVS gene has been transcribed into RNA, ribosomes can translate the MAVS protein from two different sites.[7] The initial translation site generates the full-length MAVS protein. The alternative translation site generates a shorter protein, termed as “miniMAVS” or short-MAVS (sMAVS).[7] sMAVS is a 398 amino acid MAVS protein that lacks the CARD domain. This is significant because the CARD domain is where two cytosolic proteins bind to activate MAVS, signaling that there is a virus present in the cell.[7]
Function
Double stranded RNA viruses are recognized by either the transmembrane toll-like receptor 3 (TLR3) or by one of two cytosolic proteins, retinoic acid-inducible gene I (RIG-I)-like receptors and melanoma differentiation-associated gene 5 (MDA5).[7][8][9][10] RIG-I and MDA5 differ in the viral RNA that they recognize, but they share many structural features, including the N-terminal CARD that allows them to bind to MAVS.[7] MAVS activation leads to the increased levels of pro-inflammatory cytokines via activation of transcription factors, nuclear factor kB (NF-kB), interferon regulatory factor 1 (IRF1), and interferon regulatory factor 3 (IRF3).[7][8][9] NFkB, IRF1, and IRF3 are transcription factors and play critical roles in the production of cytokines.
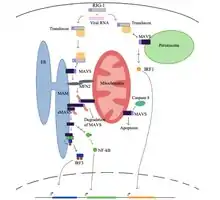
At a resting state for the cell, a protein called mitofusin 2 (MFN2) is known to interact with MAVS, preventing MAVS from binding to the cytosolic proteins, such as RIG-I and MDA5.[7][8] Upon recognition of the virus in the cytosol, mitochondria-associated ER membranes (MAM) and mitochondria will become physically tethered by MFN2 and RIG-I binds to a second RIG-I protein to form a protein complex.[7][8][9] This complex binds to TRIM25 and molecular chaperone 14-3-3e to form a complex termed “translocon”.[7][8][9][10] The translocon travels to the mitochondria where it binds to the CARD region on MAVS, leading to activation of MAVS.[7][8][9][10] Subsequently, MAVS proteins bind to each other through the CARD and TM domain to recruit several downstream signaling factors to form the MAVS signaling complex.[7][8] The formation of this MAVS signaling complex is aided by augmented levels of mitochondrial reactive oxygen species (mROS), independent of the RNA sensing.[8][9] The MAVS signaling complex interacts with TANK binding kinase 1 and/or protein kinases IKKA (CHUK) and IKKB (IKBKB), which leads to the phosphorylation and nuclear translocation of IRF3.[7] Although MAVS signal transduction and regulation is not fully understood, activated MAVS proteins in the mitochondria, ER, and peroxisome are needed to maximize the antiviral innate immune response.
MAVS protein induces apoptosis in host virally infected cells by interacting with a protease called caspase 8.[7] Activation of apoptosis by caspase 8 is independent of the Bax/Bak apoptotic pathway, the main pathway of apoptosis in cells.[7]
Viral evasion
Certain viruses, such as human cytomegalovirus (HCMV) and hepatitis C (HCV), have adapted to suppress the function of MAVS in the antiviral innate immune response, aiding in viral replication.[7][11] HCMV impairs MAVS through the viral mitochondria-localized inhibitor of apoptosis protein (vMIA), thus reducing the pro-inflammatory cytokine response.[11] vMIA also localizes to the peroxisome where it interacts with cytoplasmic chaperone protein Pex19, disabling the transport machinery of peroxisomal membrane proteins.[11] The HCV NS3-NS4A strain inactivates MAVS signaling by cleaving the MAVS protein directly upstream of MAVS membrane-targeting domain in the MAM and peroxisome, preventing MAVS downstream signaling.[7]
Regulation
The expression and function of MAVS are regulated at the transcriptional, posttranscriptional, and posttranslational level. At the transcriptional level, the reactive oxygen species (ROS) generated during antiviral response acts as a negative regulator.[7][8][9] MAVS, additionally, encodes a number of splice variants that have been proposed to regulate MAVS. At the post-transcriptional level, there are two translational sites present on MAVS that can generate two proteins of MAVS. The alternative translation site resides upstream, resulting in expression of sMAVS.[7][8][9] At the translational level, proteins such as a family of ubiquitin E3 ligase regulate MAVS activity.[7][8][9]
References
- GRCh38: Ensembl release 89: ENSG00000088888 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000037523 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Vazquez C, Beachboard DC, Horner SM (2017). "Methods to Visualize MAVS Subcellular Localization". Innate Antiviral Immunity. Methods in Molecular Biology. Vol. 1656. pp. 131–142. doi:10.1007/978-1-4939-7237-1_7. ISBN 978-1-4939-7236-4. PMC 6103534. PMID 28808966.
- Seth RB, Sun L, Ea CK, Chen ZJ (September 2005). "Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3". Cell. 122 (5): 669–682. doi:10.1016/j.cell.2005.08.012. PMID 16125763. S2CID 11104354.
- Vazquez C, Horner SM (July 2015). "MAVS Coordination of Antiviral Innate Immunity". Journal of Virology. 89 (14): 6974–6977. doi:10.1128/JVI.01918-14. PMC 4473567. PMID 25948741.
- Mohanty A, Tiwari-Pandey R, Pandey NR (September 2019). "Mitochondria: the indispensable players in innate immunity and guardians of the inflammatory response". Journal of Cell Communication and Signaling. 13 (3): 303–318. doi:10.1007/s12079-019-00507-9. PMC 6732146. PMID 30719617.
- Jacobs JL, Coyne CB (December 2013). "Mechanisms of MAVS regulation at the mitochondrial membrane". Journal of Molecular Biology. 425 (24): 5009–5019. doi:10.1016/j.jmb.2013.10.007. PMC 4562275. PMID 24120683.
- Jiang QX (2019). "Structural Variability in the RLR-MAVS Pathway and Sensitive Detection of Viral RNAs". Medicinal Chemistry. 15 (5): 443–458. doi:10.2174/1573406415666181219101613. PMC 6858087. PMID 30569868.
- Ashley CL, Abendroth A, McSharry BP, Slobedman B (2019). "Interferon-Independent Innate Responses to Cytomegalovirus". Frontiers in Immunology. 10: 2751. doi:10.3389/fimmu.2019.02751. PMC 6917592. PMID 31921100.
Further reading
- Nagase T, Ishikawa K, Kikuno R, Hirosawa M, Nomura N, Ohara O (October 1999). "Prediction of the coding sequences of unidentified human genes. XV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Research. 6 (5): 337–345. doi:10.1093/dnares/6.5.337. PMID 10574462.
- Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, Nagano Y, et al. (May 2003). "Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways". Oncogene. 22 (21): 3307–3318. doi:10.1038/sj.onc.1206406. PMID 12761501.
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. (October 2005). "IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction". Nature Immunology. 6 (10): 981–988. doi:10.1038/ni1243. PMID 16127453. S2CID 31479259.
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J (October 2005). "Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus" (PDF). Nature. 437 (7062): 1167–1172. doi:10.1038/nature04193. PMID 16177806. S2CID 4391603.
- Li XD, Sun L, Seth RB, Pineda G, Chen ZJ (December 2005). "Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity". Proceedings of the National Academy of Sciences of the United States of America. 102 (49): 17717–17722. doi:10.1073/pnas.0508531102. PMC 1308909. PMID 16301520.
- Oh JH, Yang JO, Hahn Y, Kim MR, Byun SS, Jeon YJ, et al. (December 2005). "Transcriptome analysis of human gastric cancer". Mammalian Genome. 16 (12): 942–954. doi:10.1007/s00335-005-0075-2. PMID 16341674. S2CID 69278.
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, et al. (April 2006). "Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection". Proceedings of the National Academy of Sciences of the United States of America. 103 (15): 6001–6006. doi:10.1073/pnas.0601523103. PMC 1458687. PMID 16585524.
- Cheng G, Zhong J, Chisari FV (May 2006). "Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms". Proceedings of the National Academy of Sciences of the United States of America. 103 (22): 8499–8504. doi:10.1073/pnas.0602957103. PMC 1482521. PMID 16707574.
- Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, et al. (June 2006). "Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage". Journal of Virology. 80 (12): 6072–6083. doi:10.1128/JVI.02495-05. PMC 1472616. PMID 16731946.
- Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, et al. (July 2006). "Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif". The EMBO Journal. 25 (14): 3257–3263. doi:10.1038/sj.emboj.7601220. PMC 1523175. PMID 16858409.
- Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP (October 2006). "A probability-based approach for high-throughput protein phosphorylation analysis and site localization". Nature Biotechnology. 24 (10): 1285–1292. doi:10.1038/nbt1240. PMID 16964243. S2CID 14294292.
- Opitz B, Vinzing M, van Laak V, Schmeck B, Heine G, Günther S, et al. (November 2006). "Legionella pneumophila induces IFNbeta in lung epithelial cells via IPS-1 and IRF3, which also control bacterial replication". The Journal of Biological Chemistry. 281 (47): 36173–36179. doi:10.1074/jbc.M604638200. PMID 16984921.
- Chen Z, Benureau Y, Rijnbrand R, Yi J, Wang T, Warter L, et al. (January 2007). "GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS". Journal of Virology. 81 (2): 964–976. doi:10.1128/JVI.02076-06. PMC 1797450. PMID 17093192.
- Hirata Y, Broquet AH, Menchén L, Kagnoff MF (October 2007). "Activation of innate immune defense mechanisms by signaling through RIG-I/IPS-1 in intestinal epithelial cells". Journal of Immunology. 179 (8): 5425–5432. doi:10.4049/jimmunol.179.8.5425. PMID 17911629.
- Zeng W, Xu M, Liu S, Sun L, Chen ZJ (October 2009). "Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3". Molecular Cell. 36 (2): 315–325. doi:10.1016/j.molcel.2009.09.037. PMC 2779157. PMID 19854139.
- Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, et al. (August 2013). "MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades". eLife. 2 (e00785): e00785. doi:10.7554/eLife.00785. PMC 3743401. PMID 23951545.
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. (March 2015). "Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation". Science. 347 (6227): aaa2630. doi:10.1126/science.aaa2630. PMID 25636800.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.