Oryza sativa
Oryza sativa, also known as rice, is the plant species most commonly referred to in English as rice. It is the type of farmed rice whose cultivars are most common globally, and was first domesticated in the Yangtze River basin in China 13,500 to 8,200 years ago.[2][3][4][5]
| Oryza sativa | |
|---|---|
_by_Augustus_Binu.jpg.webp) | |
| Mature seed heads | |
 | |
| Inflorescence | |
| Scientific classification | |
| Kingdom: | Plantae |
| Clade: | Tracheophytes |
| Clade: | Angiosperms |
| Clade: | Monocots |
| Clade: | Commelinids |
| Order: | Poales |
| Family: | Poaceae |
| Genus: | Oryza |
| Species: | O. sativa |
| Binomial name | |
| Oryza sativa | |
| Synonyms[1] | |
|
List
| |
Oryza sativa belongs to the genus Oryza of the grass family Poaceae. With a genome consisting of 430 Mbp across 12 chromosomes, it is renowned for being easy to genetically modify and is a model organism for the botany of cereals.
Botany
The species has an erect and stout or slender stalk stem that grows 80−120 centimetres tall, with a smooth surface. The leaf is lanceolate, 15−30 cm long, and grows from a ligule 10−20 millimetres long.[6]
Classification
Oryza sativa contains two major subspecies: the sticky, short-grained japonica or sinica variety, and the nonsticky, long-grained indica [zh] [ja] rice variety. Japonica was domesticated in the Yangtze Valley 9–6,000 years ago,[7] and its varieties can be cultivated in dry fields (it is cultivated mainly submerged in Japan), in temperate East Asia, upland areas of Southeast Asia, and high elevations in South Asia, while indica was domesticated around the Ganges 8,500-4,500 years ago,[7] and its varieties are mainly lowland rices, grown mostly submerged, throughout tropical Asia. Rice occurs in a variety of colors, including white, brown, black, purple, and red rices.[8][9] Black rice (also known as purple rice) is a range of rice types, some of which are glutinous rice. Varieties include Indonesian black rice and Thai jasmine black rice.
A third subspecies, which is broad-grained and thrives under tropical conditions, was identified based on morphology and initially called javanica, but is now known as tropical japonica. Examples of this variety include the medium-grain 'Tinawon' and 'Unoy' cultivars, which are grown in the high-elevation rice terraces of the Cordillera Mountains of northern Luzon, Philippines.[10]
Glaszmann (1987) used isozymes to sort O. sativa into six groups: japonica, aromatic, indica, aus, rayada, and ashina.[11]
Garris et al. (2004) used simple sequence repeats to sort O. sativa into five groups: temperate japonica, tropical japonica and aromatic comprise the japonica varieties, while indica and aus comprise the indica varieties.[12]
Nomenclature and taxonomy
Rice has been cultivated since ancient times and oryza[13] is a classical Latin word for rice while sativa[14] means "cultivated".
Genetics
SPL14/LOC4345998 is a gene that regulates the overall architecture/growth habit of the plant. Some of its epialleles increase rice yield.[15] An accurate and usable Simple Sequence Repeat marker set was developed and used to generate a high-density map in McCouch et al., 2002.[16] A multiplex high-throughput marker assisted selection system has been developed by Masouleh et al., 2009 but as with other crop HTMAS systems has proven difficult to customize, costly (both directly and for the equipment), and inflexible.[16] Other molecular breeding tools have produced results, producing blast resistant cultivars.[17][18][16] Xu et al., 2014 uses a DNA microarray to substantially advance understanding of hybrid vigor in rice, Takagi et al., 2013 uses QTL sequencing to elucidate seedling vigor, and Yano et al., 2016 performs a genome wide association study (GWAS) by whole genome sequencing (WGS) to investigate various agronomic traits.[16] (Because the correspondence between genotype and phenotype is more easily understood in rice, translation of results from rice to other non-models may require more work. For example, grain size and grain weight in wheat were elucidated in this way by Valluru et al., 2014.)[16]
Affymetrix offers a 44 thousand pot microarray, a 50 thousand, and a one million, and Illumina has a six thousand and a 50 thousand, all of which have performed well and are commonly used.[16] Rice is one of the earliest uses and validation models for the semi-thermal asymmetric reverse PCR (STARP) method developed in Long et al., 2016.[16] The putative homolog for spindle and kinetochore-associated protein 1 – OsSka1 – is localized to XP_478114 by Hanisch et al., 2006.[19]
Resistance to Magnaporthe grisea is provided by various resistance genes including Pi1, Pi54, and Pita.[20]
O. sativa has a large number of insect resistance genes specifically for the Brown planthopper.[21] As of 2022 15 R genes among these have been cloned and characterized including Tamura et al., 2014's[22] discovery of Bph2[23][21] Guo et al., 2018's discovery of Bph6,[21] Zhao et al., 2016's discovery of Bph9,[21] Du et al., 2009's discovery of Bph14,[21] and Ji et al., 2016's[24] discovery of Bph18.[23][21]
In total 641 copy number variations are known, the combination of results of Ma and Bennetzen 2004 and Yu et al., 2011.[16] Exome capture often reveals new single nucleotide polymorphisms in rice, due to its large genome and high degree of DNA repetition.[16] There have been two major results of this type, Saintenac et al., 2011 and Henry et al., 2014.[16]
Both abscisic acid and salicylic acid are employed by O. sativa in its regulation of its own immune responses.[25] Jiang et al., 2010 finds SA broadly upregulates and ABA broadly downregulates immunity to M. grisea, and success depends on the balance between their levels.[25]
Breeding

While most rice is bred for crop quality and productivity, there are varieties selected for characteristics such as texture, smell, and firmness. There are four major categories of rice worldwide: indica, japonica, aromatic and glutinous. The different varieties of rice are not considered interchangeable, either in food preparation or agriculture, so as a result, each major variety is a completely separate market from other varieties. It is common for one variety of rice to rise in price while another one drops in price.
Rice cultivars also fall into groups according to environmental conditions, season of planting, and season of harvest, called ecotypes. Some major groups are the Japan-type (grown in Japan), "buly" and "tjereh" types (Indonesia); sali (or aman—main winter crop), ahu (also aush or ghariya, summer), and boro (spring) (Bengal and Assam). Cultivars exist that are adapted to deep flooding, and these are generally called "floating rice".A triple introgression of resistance genes against Magnaporthe grisea—and actual field resistance—have been demonstrated by Khan et al., 2018.[20] This is a marker-assisted backcross of Pi1, Pi54, and Pita into an aromatic cultivar using SSR- and STS-markers.[20] Pi21 is protein gene.[26] An allele pi21 confers broad-spectrum non-race-specific blast resistance against many strain.[26]
Gallery
 Water buffalo ploughing, Java
Water buffalo ploughing, Java Jumli Marshi, brown rice from Nepal
Jumli Marshi, brown rice from Nepal Traditional rice of Niyamgiri Hills, India
Traditional rice of Niyamgiri Hills, India From Chhattisgarh
From Chhattisgarh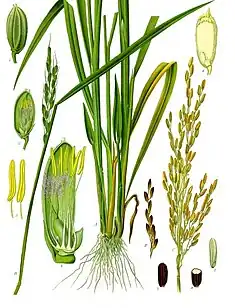
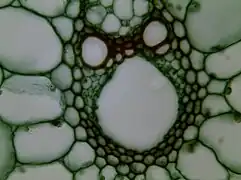 Stem cross section magnified 400 times
Stem cross section magnified 400 times
See also
References
- "Oryza sativa L." Plants of the World Online. Board of Trustees of the Royal Botanic Gardens, Kew. 2017. Retrieved December 21, 2020.
- Normile, Dennis (1997). "Yangtze seen as earliest rice site". Science. 275 (5298): 309–310. doi:10.1126/science.275.5298.309. S2CID 140691699.
- Vaughan, DA; Lu, B; Tomooka, N (2008). "The evolving story of rice evolution". Plant Science. 174 (4): 394–408. doi:10.1016/j.plantsci.2008.01.016.
- Harris, David R. (1996). The Origins and Spread of Agriculture and Pastoralism in Eurasia. Psychology Press. p. 565. ISBN 978-1-85728-538-3.
- Zhang, Jianping; Lu, Houyuan; Gu, Wanfa; Wu, Naiqin; Zhou, Kunshu; Hu, Yayi; Xin, Yingjun; Wang, Can; Kashkush, Khalil (December 17, 2012). "Early Mixed Farming of Millet and Rice 7800 Years Ago in the Middle Yellow River Region, China". PLOS ONE. 7 (12): e52146. Bibcode:2012PLoSO...752146Z. doi:10.1371/journal.pone.0052146. PMC 3524165. PMID 23284907.
- Catindig, J.L.A.; Lubigan, R.T.; Johnson, D. (n.d.). "Oryza sativa". Rice Knowledge Bank. International Rice Research Institute. Retrieved June 29, 2023.
- Purugganan, Michael D.; Fuller, Dorian Q. (2009). "The nature of selection during plant domestication". Nature. Nature Research. 457 (7231): 843–848. Bibcode:2009Natur.457..843P. doi:10.1038/nature07895. ISSN 0028-0836. PMID 19212403. S2CID 205216444.
- Oka (1988)
- Mohammadi Shad, Z; Atungulu, G. (March 2019). "Post-harvest kernel discoloration and fungi activity in long-grain hybrid, pureline and medium-grain rice cultivars as influenced by storage environment and antifungal treatment". Journal of Stored Products Research. 81: 91–99. doi:10.1016/j.jspr.2019.02.002. S2CID 92050510.
- CECAP, PhilRice and IIRR. 2000. "Highland Rice Production in the Philippine Cordillera."
- Glaszmann, J. C. (May 1987). "Isozymes and classification of Asian rice varieties". Theoretical and Applied Genetics. 74 (1): 21–30. doi:10.1007/BF00290078. PMID 24241451. S2CID 22829122.
- Garris; Tai, TH; Coburn, J; Kresovich, S; McCouch, S (2004). "Genetic structure and diversity in Oryza sativa L." Genetics. 169 (3): 1631–8. doi:10.1534/genetics.104.035642. PMC 1449546. PMID 15654106.
- "oryza". Merriam-Webster.com Dictionary.
-
- "sativa". Lexico UK English Dictionary. Oxford University Press. n.d.
- "sativa". Merriam-Webster.com Dictionary.
- Stange, Madlen; Barrett, Rowan D. H.; Hendry, Andrew P. (February 2021). "The importance of genomic variation for biodiversity, ecosystems and people". Nature Reviews Genetics. Nature Portfolio. 22 (2): 89–105. doi:10.1038/s41576-020-00288-7. ISSN 1471-0056. PMID 33067582. S2CID 223559538. MS ORCID 0000-0002-4559-2535). (RDHB ORCID 0000-0003-3044-2531).
- Rasheed, Awais; Hao, Yuanfeng; Xia, Xianchun; Khan, Awais; Xu, Yunbi; Varshney, Rajeev K.; He, Zhonghu (2017). "Crop Breeding Chips and Genotyping Platforms: Progress, Challenges, and Perspectives". Molecular Plant. Elsevier. 10 (8): 1047–1064. doi:10.1016/j.molp.2017.06.008. ISSN 1674-2052. PMID 28669791. S2CID 33780984. Chinese Academy of Sciences+Chinese Society for Plant Biology+Shanghai Institutes for Biological Sciences.
- Miah, G.; Rafii, M. Y.; Ismail, M. R.; Puteh, A. B.; Rahim, H. A.; Asfaliza, R.; Latif, M. A. (November 27, 2012). "Blast resistance in rice: a review of conventional breeding to molecular approaches" (PDF). Molecular Biology Reports. Springer Science+Business Media. 40 (3): 2369–2388. doi:10.1007/s11033-012-2318-0. ISSN 0301-4851. PMID 23184051. S2CID 8922855.
- Rao, Yuchun; Li, Yuanyuan; Qian, Qian (January 19, 2014). "Recent progress on molecular breeding of rice in China". Plant Cell Reports. Springer Science+Business Media. 33 (4): 551–564. doi:10.1007/s00299-013-1551-x. ISSN 0721-7714. PMC 3976512. PMID 24442397.
- Cheeseman, Iain M.; Desai, Arshad (2008). "Molecular architecture of the kinetochore–microtubule interface". Nature Reviews Molecular Cell Biology. Nature Portfolio. 9 (1): 33–46. doi:10.1038/nrm2310. ISSN 1471-0072. PMID 18097444. S2CID 34121605.
- Mehta, Sahil; Singh, Baljinder; Dhakate, Priyanka; Rahman, Mehzabin; Islam, Muhammad Aminul (2019). "5 Rice, Marker-Assisted Breeding, and Disease Resistance". In Wani, Shabir Hussain (ed.). Disease Resistance in Crop Plants : Molecular, Genetic and Genomic Perspectives. Cham, Switzerland: Springer. pp. 83–112/xii+307. ISBN 978-3-030-20727-4. OCLC 1110184027.
- Wang, Changsheng; Han, Bin (2022). "Twenty years of rice genomics research: From sequencing and functional genomics to quantitative genomics". Molecular Plant. Cell Press. 15 (4): 593–619. doi:10.1016/j.molp.2022.03.009. ISSN 1674-2052. PMID 35331914. S2CID 247603925.
- Tamura, Yasumori; Hattori, Makoto; Yoshioka, Hirofumi; Yoshioka, Miki; Takahashi, Akira; Wu, Jianzhong; Sentoku, Naoki; Yasui, Hideshi (July 29, 2014). "Map-based Cloning and Characterization of a Brown Planthopper Resistance Gene BPH26 from Oryza sativa L. ssp. indica Cultivar ADR52". Scientific Reports. Nature Portfolio. 4 (1): 5872. Bibcode:2014NatSR...4E5872T. doi:10.1038/srep05872. ISSN 2045-2322. PMC 5376202. PMID 25076167.
- Jing, Shengli; Zhao, Yan; Du, Bo; Chen, Rongzhi; Zhu, Lili; He, Guangcun (2017). "Genomics of interaction between the brown planthopper and rice". Current Opinion in Insect Science. Elsevier. 19: 82–87. doi:10.1016/j.cois.2017.03.005. ISSN 2214-5745. PMID 28521948.
- Ji, Hyeonso; Kim, Sung-Ryul; Kim, Yul-Ho; Suh, Jung-Pil; Park, Hyang-Mi; Sreenivasulu, Nese; Misra, Gopal; Kim, Suk-Man; Hechanova, Sherry Lou; Kim, Hakbum; Lee, Gang-Seob; Yoon, Ung-Han; Kim, Tae-Ho; Lim, Hyemin; Suh, Suk-Chul; Yang, Jungil; An, Gynheung; Jena, Kshirod K. (September 29, 2016). "Map-based Cloning and Characterization of the BPH18 Gene from Wild Rice Conferring Resistance to Brown Planthopper (BPH) Insect Pest". Scientific Reports. Nature Portfolio. 6 (1): 34376. Bibcode:2016NatSR...634376J. doi:10.1038/srep34376. ISSN 2045-2322. PMC 5041133. PMID 27682162. S2CID 1034311.
-
- Pieterse, Corné M.J.; Van der Does, Dieuwertje; Zamioudis, Christos; Leon-Reyes, Antonio; Van Wees, Saskia C.M. (November 10, 2012). "Hormonal Modulation of Plant Immunity". Annual Review of Cell and Developmental Biology. Annual Reviews. 28 (1): 489–521. doi:10.1146/annurev-cellbio-092910-154055. hdl:1874/274421. ISSN 1081-0706. PMID 22559264. S2CID 18180536.
- Atkinson, Nicky J.; Urwin, Peter E. (March 30, 2012). "The interaction of plant biotic and abiotic stresses: from genes to the field". Journal of Experimental Botany. Oxford University Press. 63 (10): 3523–3543. doi:10.1093/jxb/ers100. ISSN 0022-0957. PMID 22467407. S2CID 205195661.
- Liu, Wende; Liu, Jinling; Triplett, Lindsay; Leach, Jan E.; Wang, Guo-Liang (August 4, 2014). "Novel Insights into Rice Innate Immunity Against Bacterial and Fungal Pathogens". Annual Review of Phytopathology. Annual Reviews. 52 (1): 213–241. doi:10.1146/annurev-phyto-102313-045926. ISSN 0066-4286. PMID 21380629. S2CID 9244874.
- Li, Wei; Deng, Yiwen; Ning, Yuese; He, Zuhua; Wang, Guo-Liang (2020). "Exploiting Broad-Spectrum Disease Resistance in Crops: From Molecular Dissection to Breeding". Annual Review of Plant Biology. Annual Reviews. 71 (1): 575–603. doi:10.1146/annurev-arplant-010720-022215. ISSN 1543-5008. PMID 32197052. S2CID 214600762.
