pUC19
pUC19 is one of a series of plasmid cloning vectors created by Joachim Messing and co-workers.[1] The designation "pUC" is derived from the classical "p" prefix (denoting "plasmid") and the abbreviation for the University of California, where early work on the plasmid series had been conducted.[2] It is a circular double stranded DNA and has 2686 base pairs.[3] pUC19 is one of the most widely used vector molecules as the recombinants, or the cells into which foreign DNA has been introduced, can be easily distinguished from the non-recombinants based on color differences of colonies on growth media. pUC18 is similar to pUC19, but the MCS region is reversed.

Components
Notably, it has an N-terminal fragment of β-galactosidase (lacZ) gene of E. coli.[4] The multiple cloning site (MCS) region is split into codons 6-7 of the lacZ gene, providing for many restriction endonucleases restriction sites.[5] In addition to β-galactosidase, pUC19 also encodes for an ampicillin resistance gene (ampR), via a β-lactamase enzyme that functions by degrading ampicillin and reducing its toxicity to the host.[6]
The ori site, or origin of replication, is derived from the plasmid pMB1. pUC19 is small but has a high copy number. The high copy number is a result of the lack of the rop gene and a single point mutation in the ori of pMB1.[7] The lacZ gene codes for β-galactosidase. The recognition sites for HindIII, SphI, PstI, SalI, XbaI, BamHI, SmaI, KpnI, SacI and EcoRI restriction enzymes have been derived from the vector M13mp19.[8]
Function
This plasmid is introduced into a bacterial cell by a process called "transformation", where it can multiply and express itself. However, due to the presence of MCS and several restriction sites, a foreign piece of DNA of choice can be introduced into it by inserting it into place in MCS region. The cells which have taken up the plasmid can be differentiated from cells which have not taken up the plasmid by growing it on media with ampicillin. Only the cells with the plasmid containing the ampicillin resistance (ampR) gene will survive. Furthermore, the transformed cells containing the plasmid with the gene of interest can be distinguished from cells with the plasmid but without the gene of interest, just by looking at the color of the colony they make on agar media supplemented with IPTG and X-gal. Recombinants are white, whereas non-recombinants are blue.
Mechanism
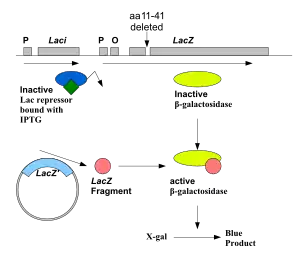
The lac Z fragment, whose synthesis can be induced by IPTG, is capable of intra-allelic complementation with a defective form of β-galactosidase enzyme encoded by host chromosome (mutation lacZDM15 in E. coli JM109, DH5α and XL1-Blue strains).[9] In the presence of IPTG in growth medium, bacteria synthesise both fragments of the enzyme. Both the fragments can together hydrolyse X-gal (5-bromo-4-chloro-3-indolyl- beta-D-galactopyranoside) and form blue colonies when grown on media where it is supplemented.
Insertion of foreign DNA into the MCS located within the lac Z gene causes insertional inactivation of this gene at the N-terminal fragment of beta-galactosidase and abolishes intra-allelic complementation. Thus bacteria carrying recombinant plasmids in the MCS cannot hydrolyse X-gal, giving rise to white colonies, which can be distinguished on culture media from non-recombinant cells, which are blue.[10]
Therefore, the media used should contain ampicillin, IPTG, and X-gal.
Use in research
Due to its extensive use as a cloning vector in research and industry, pUC19 is frequently used in research as a model plasmid.[11] For example, biophysical studies on its naturally supercoiled state have determined its radius of gyration to be 65.6 nm and its Stokes radius to be 43.6 nm.
References
- Yanisch-Perron, C.; Vieira, J.; Messing, J. (1985). "Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors". Gene. 33 (1): 103–119. doi:10.1016/0378-1119(85)90120-9. PMID 2985470.
- Vieira, J.; Messing, J. (1982). "The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers". Gene. 19 (3): 259–268. doi:10.1016/0378-1119(82)90015-4. PMID 6295879.
- pUC19 description & restriction map
- Louro, Ricardo O.; Crichton, Robert R. (2013). Practical approaches to biological inorganic chemistry. Amsterdam, Oxford: Elsevier. p. 279. ISBN 9780444563590. Retrieved April 7, 2014.
- Louro, Ricardo O.; Crichton, Robert R. (2013). Practical approaches to biological inorganic chemistry. Amsterdam, Oxford: Elsevier. p. 279. ISBN 9780444563590. Retrieved April 7, 2014.
- Wang, Nam Sun. "Summary of Sites on pUC19". Department of Chemical & Biomolecular Engineering University of Maryland. Retrieved 27 January 2017.
- Saha; et al. (2004). "A naturally occurring point mutation in the 13-mer R repeat affects the oriC function of the large chromosome of Vibrio cholerae O1 classical biotype". Archives of Microbiology. 182 (5): 421–427. doi:10.1007/s00203-004-0708-y. PMID 15375645. S2CID 28286917.
- Mooreland; et al. (2013). Advanced Biomolecular Genetics. Kleiske Publishing. pp. 889–932.
- Louro, Ricardo O.; Crichton, Robert R. (2013). Practical approaches to biological inorganic chemistry. Amsterdam, Oxford: Elsevier. p. 279. ISBN 9780444563590. Retrieved April 7, 2014.
- Pasternak, Jack J. (2005). An Introduction to Human Molecular Genetics, Second Edition. Wiley-IEEE. p. 117. ISBN 978-0-471-71917-5.
- Störkle, Dominic (5 September 2007). "Complex Formation of DNA with Oppositely Charged Polyelectrolytes of Different Chain Topology: Cylindrical Brushes and Dendrimers". Macromolecules. 40 (22): 7998–8006. Bibcode:2007MaMol..40.7998S. doi:10.1021/ma0711689.