Phosphatidic acid
Phosphatidic acids are anionic phospholipids important to cell signaling and direct activation of lipid-gated ion channels. Hydrolysis of phosphatidic acid gives rise to one molecule each of glycerol and phosphoric acid and two molecules of fatty acids. They constitute about 0.25% of phospholipids in the bilayer.[1]
Structure
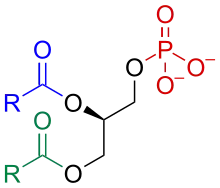
Phosphatidic acid consists of a glycerol backbone, with, in general, a saturated fatty acid bonded to carbon-1, an unsaturated fatty acid bonded to carbon-2, and a phosphate group bonded to carbon-3.[2][3]
Formation and degradation
Besides de novo synthesis, PA can be formed in three ways:
- By phospholipase D (PLD), via the hydrolysis of the P-O bond of phosphatidylcholine (PC) to produce PA and choline.[4]
- By the phosphorylation of diacylglycerol (DAG) by DAG kinase (DAGK).
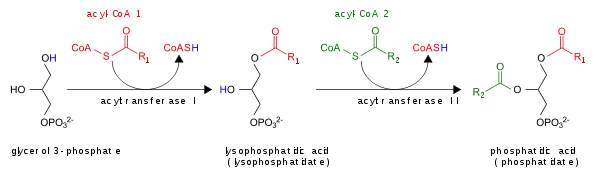
- By the acylation of lysophosphatidic acid by lysoPA-acyltransferase (LPAAT); this is the most common pathway.[5]
The glycerol 3-phosphate pathway for de novo synthesis of PA is shown here:
In addition, PA can be converted into DAG by lipid phosphate phosphohydrolases (LPPs)[6][7] or into lyso-PA by phospholipase A (PLA).
Roles in the cell
The role of PA in the cell can be divided into three categories:
- PA is the precursor for the biosynthesis of many other lipids.
- The physical properties of PA influence membrane curvature.
- PA acts as a signaling lipid, recruiting cytosolic proteins to appropriate membranes (e.g., sphingosine kinase 1[8]).
- PA plays very important role in phototransduction in Drosophila.[9]
- PA is a lipid ligand that gates ion channels.[10] See also lipid-gated ion channels.
The first three roles are not mutually exclusive. For example, PA may be involved in vesicle formation by promoting membrane curvature and by recruiting the proteins to carry out the much more energetically unfavourable task of neck formation and pinching.
Roles in biosynthesis
PA is a vital cell lipid that acts as a biosynthetic precursor for the formation (directly or indirectly) of all acylglycerol lipids in the cell.[11]
In mammalian and yeast cells, two different pathways are known for the de novo synthesis of PA, the glycerol 3-phosphate pathway or the dihydroxyacetone phosphate pathway. In bacteria, only the former pathway is present, and mutations that block this pathway are lethal, demonstrating the importance of PA. In mammalian and yeast cells, where the enzymes in these pathways are redundant, mutation of any one enzyme is not lethal. However, it is worth noting that in vitro, the various acyltransferases exhibit different substrate specificities with respect to the acyl-CoAs that are incorporated into PA. Different acyltransferases also have different intracellular distributions, such as the endoplasmic reticulum (ER), the mitochondria or peroxisomes, and local concentrations of activated fatty acids. This suggests that the various acyltransferases present in mammalian and yeast cells may be responsible for producing different pools of PA.[11]
The conversion of PA into diacylglycerol (DAG) by LPPs is the commitment step for the production of phosphatidylcholine (PC), phosphatidylethanolamine (PE) and phosphatidylserine (PS). In addition, DAG is also converted into CDP-DAG, which is a precursor for phosphatidylglycerol (PG), phosphatidylinositol (PI) and phosphoinositides (PIP, PIP2, PIP3).[11]
PA concentrations are maintained at extremely low levels in the cell by the activity of potent LPPs.[6] These convert PA into DAG very rapidly and, because DAG is the precursor for so many other lipids, it too is soon metabolised into other membrane lipids. This means that any upregulation in PA production can be matched, over time, with a corresponding upregulation in LPPs and in DAG metabolising enzymes.
PA is, therefore, essential for lipid synthesis and cell survival, yet, under normal conditions, is maintained at very low levels in the cell.
Biophysical properties
PA is a unique phospholipid in that it has a small highly charged head group that is very close to the glycerol backbone. PA is known to play roles in both vesicle fission[12] and fusion,[13] and these roles may relate to the biophysical properties of PA.
At sites of membrane budding or fusion, the membrane becomes or is highly curved. A major event in the budding of vesicles, such as transport carriers from the Golgi, is the creation and subsequent narrowing of the membrane neck. Studies have suggested that this process may be lipid-driven, and have postulated a central role for DAG due to its, likewise, unique molecular shape. The presence of two acyl chains but no headgroup results in a large negative curvature in membranes.[14]
The LPAAT BARS-50 has also been implicated in budding from the Golgi.[12] This suggests that the conversion of lysoPA into PA might affect membrane curvature. LPAAT activity doubles the number of acyl chains, greatly increasing the cross-sectional area of the lipid that lies ‘within’ the membrane while the surface headgroup remains unchanged. This can result in a more negative membrane curvature. Researchers from Utrecht University have looked at the effect of lysoPA versus PA on membrane curvature by measuring the effect these have on the transition temperature of PE from lipid bilayers to nonlamellar phases using 31P-NMR.[15] The curvature induced by these lipids was shown to be dependent not only on the structure of lysoPA versus PA but also on dynamic properties, such as the hydration of head groups and inter- and intramolecular interactions. For instance, Ca2+ may interact with two PAs to form a neutral but highly curved complex. The neutralisation of the otherwise repulsive charges of the headgroups and the absence of any steric hindrance enables strong intermolecular interactions between the acyl chains, resulting in PA-rich microdomains. Thus in vitro, physiological changes in pH, temperature, and cation concentrations have strong effects on the membrane curvature induced by PA and lysoPA.[15] The interconversion of lysoPA, PA, and DAG – and changes in pH and cation concentration – can cause membrane bending and destabilisation, playing a direct role in membrane fission simply by virtue of their biophysical properties. However, though PA and lysoPA have been shown to affect membrane curvature in vitro; their role in vivo is unclear.
The roles of lysoPA, PA, and DAG in promoting membrane curvature do not preclude a role in recruiting proteins to the membrane. For instance, the Ca2+ requirement for the fusion of complex liposomes is not greatly affected by the addition of annexin I, though it is reduced by PLD. However, with annexin I and PLD, the extent of fusion is greatly enhanced, and the Ca2+ requirement is reduced almost 1000-fold to near physiological levels.[13]
Thus the metabolic, biophysical, recruitment, and signaling roles of PA may be interrelated.
Role in signaling
PA is kept low in the bulk of the membrane in order to transiently burst and signal locally in high concentration.[16] For example TREK-1 channels are activated by local association with PLD and production of PA.[17] The dissociation constant of PA for TREK-1 is approximately 10 micromolar.[18] The relatively weak binding combined with a low concentration of PA in the membrane allows the channel to turn off. The local high concentration for activation suggests at least some restrictions in local lipid diffusion. The bulk low concentration of PA combined with high local bursts is the opposite of PIP2 signaling. PIP2 is kept relatively high in the membrane and then transiently hydrolized near a protein in order to transiently reduce PIP2 signaling.[19] PA signaling mirrors PIP2 signaling in that the bulk concentration of signaling lipid need not change to exert a potent local effect on a target protein.
As described above, PLD hydrolyzes PC to form PA and choline. Because choline is very abundant in the cell, PLD activity does not significantly affect choline levels; and choline is unlikely to play any role in signaling.
The role of PLD activation in numerous signaling contexts, combined with the lack of a role for choline, suggests that PA is important in signaling. However, PA is rapidly converted to DAG, and DAG is also known to be a signaling molecule. This raises the question as to whether PA has any direct role in signaling or whether it simply acts as a precursor for DAG production.[20][21] If it is found that PA acts only as a DAG precursor, then one can raise the question as to why cells should produce DAG using two enzymes when they contain the PLC that could produce DAG in a single step.
PA produced by PLD or by DAGK can be distinguished by the addition of [γ-32P]ATP. This will show whether the phosphate group is newly derived from the kinase activity or whether it originates from the PC.[22]
Although PA and DAG are interconvertible, they do not act in the same pathways. Stimuli that activate PLD do not activate enzymes downstream of DAG, and vice versa. For example, it was shown that addition of PLD to membranes results in the production of [32P]-labeled PA and [32P]-labeled phosphoinositides.[23] The addition of DAGK inhibitors eliminates the production of [32P]-labeled PA but not the PLD-stimulated production of phosphoinositides.
It is possible that, though PA and DAG are interconvertible, separate pools of signaling and non-signaling lipids may be maintained. Studies have suggested that DAG signaling is mediated by polyunsaturated DAG, whereas PLD-derived PA is monounsaturated or saturated. Thus functional saturated/monounsaturated PA can be degraded by hydrolysing it to form non-functional saturated/monounsaturated DAG, whereas functional polyunsaturated DAG can be degraded by converting it into non-functional polyunsaturated PA.[20][24]
This model suggests that PA and DAG effectors should be able to distinguish lipids with the same headgroups but with differing acyl chains. Although some lipid-binding proteins are able to insert themselves into membranes and could hypothetically recognize the type of acyl chain or the resulting properties of the membrane, many lipid-binding proteins are cytosolic and localize to the membrane by binding only the headgroups of lipids. Perhaps the different acyl chains can affect the angle of the head-group in the membrane. If this is the case, it suggests that a PA-binding domain must not only be able to bind PA specifically but must also be able to identify those head-groups that are at the correct angle. Whatever the mechanism is, such specificity is possible. It is seen in the pig testes DAGK that is specific for polyunsaturated DAG[25] and in two rat hepatocyte LPPs that dephosphorylate different PA species with different activities.[26] Moreover, the stimulation of SK1 activity by PS in vitro was shown to vary greatly depending on whether dioleoyl (C18:1), distearoyl (C18:0), or 1-stearoyl, 2-oleoyl species of PS were used.[27] Thus it seems that, though PA and DAG are interconvertible, the different species of lipids can have different biological activities; and this may enable the two lipids to maintain separate signaling pathways.
Measurement of PA production
As PA is rapidly converted to DAG, it is very short-lived in the cell. This means that it is difficult to measure PA production and therefore to study the role of PA in the cell. However, PLD activity can be measured by the addition of primary alcohols to the cell.[28] PLD then carries out a transphosphatidylation reaction, instead of hydrolysis, producing phosphatidyl alcohols in place of PA. The phosphatidyl alcohols are metabolic dead-ends, and can be readily extracted and measured. Thus PLD activity and PA production (if not PA itself) can be measured, and, by blocking the formation of PA, the involvement of PA in cellular processes can be inferred.
Protein interactors
References
- Welti, R; Li, W; Li, M; Sang, Y; Biesiada, H; Zhou, HE; Rajashekar, CB; Williams, TD; Wang, X (30 August 2002). "Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis". The Journal of Biological Chemistry. 277 (35): 31994–2002. doi:10.1074/jbc.M205375200. PMID 12077151.
- William W. Christie. "Phosphatidic Acid, Lysophosphatidic Acid and Related Lipids". Archived from the original on 23 October 2004. Retrieved 5 November 2009.
- Schroeder, R.; London, E.; Brown, D. (December 1994). "Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior". Proceedings of the National Academy of Sciences of the United States of America. 91 (25): 12130–12134. Bibcode:1994PNAS...9112130S. doi:10.1073/pnas.91.25.12130. PMC 45390. PMID 7991596.
- Liscovitch M, Czarny M, Fiucci G, Tang X (February 2000). "Phospholipase D: molecular and cell biology of a novel gene family". Biochem. J. 345 (3): 401–15. doi:10.1042/0264-6021:3450401. PMC 1220771. PMID 10642495.
- Devlin, T. M. 2004. Bioquímica, 4ª edición. Reverté, Barcelona. ISBN 84-291-7208-4
- Brindley DN, Waggoner DW (May 1996). "Phosphatidate phosphohydrolase and signal transduction". Chem. Phys. Lipids. 80 (1–2): 45–57. doi:10.1016/0009-3084(96)02545-5. PMID 8681429.
- Brindley DN, Waggoner DW (September 1998). "Mammalian lipid phosphate phosphohydrolases". J. Biol. Chem. 273 (38): 24281–4. doi:10.1074/jbc.273.38.24281. PMID 9733709.
- Delon C, Manifava M, Wood E, et al. (October 2004). "Sphingosine kinase 1 is an intracellular effector of phosphatidic acid". J. Biol. Chem. 279 (43): 44763–74. doi:10.1074/jbc.M405771200. PMID 15310762.
- P, Raghu (August 2012). "Lipid signaling in Drosophila photoreceptors". Biochim Biophys Acta. 1821 (8): 1154–1165. doi:10.1016/j.bbalip.2012.03.008. PMID 22487656.
- Robinson, CV; Rohacs, T; Hansen, SB (September 2019). "Tools for Understanding Nanoscale Lipid Regulation of Ion Channels". Trends in Biochemical Sciences. 44 (9): 795–806. doi:10.1016/j.tibs.2019.04.001. PMC 6729126. PMID 31060927.
- Athenstaedt K, Daum G (November 1999). "Phosphatidic acid, a key intermediate in lipid metabolism". Eur. J. Biochem. 266 (1): 1–16. doi:10.1046/j.1432-1327.1999.00822.x. PMID 10542045.
- Weigert R, Silletta MG, Spanò S, et al. (November 1999). "CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid". Nature. 402 (6760): 429–33. Bibcode:1999Natur.402..429W. doi:10.1038/46587. PMID 10586885. S2CID 4423468.
- Blackwood RA, Smolen JE, Transue A, et al. (April 1997). "Phospholipase D activity facilitates Ca2+-induced aggregation and fusion of complex liposomes". Am. J. Physiol. 272 (4 Pt 1): C1279–85. doi:10.1152/ajpcell.1997.272.4.C1279. PMID 9142853.
- Shemesh T, Luini A, Malhotra V, Burger KN, Kozlov MM (December 2003). "Prefission Constriction of Golgi Tubular Carriers Driven by Local Lipid Metabolism: A Theoretical Model". Biophys. J. 85 (6): 3813–27. Bibcode:2003BpJ....85.3813S. doi:10.1016/S0006-3495(03)74796-1. PMC 1303683. PMID 14645071. Archived from the original on 2008-05-07.
- Kooijman EE, Chupin V, de Kruijff B, Burger KN (March 2003). "Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid". Traffic. 4 (3): 162–74. doi:10.1034/j.1600-0854.2003.00086.x. PMID 12656989.
- Robinson, CV; Rohacs, T; Hansen, SB (September 2019). "Tools for Understanding Nanoscale Lipid Regulation of Ion Channels". Trends in Biochemical Sciences. 44 (9): 795–806. doi:10.1016/j.tibs.2019.04.001. PMC 6729126. PMID 31060927.
- Comoglio, Y; Levitz, J; Kienzler, MA; Lesage, F; Isacoff, EY; Sandoz, G (16 September 2014). "Phospholipase D2 specifically regulates TREK potassium channels via direct interaction and local production of phosphatidic acid". Proceedings of the National Academy of Sciences of the United States of America. 111 (37): 13547–52. Bibcode:2014PNAS..11113547C. doi:10.1073/pnas.1407160111. PMC 4169921. PMID 25197053.
- Cabanos, C; Wang, M; Han, X; Hansen, SB (8 August 2017). "A Soluble Fluorescent Binding Assay Reveals PIP2 Antagonism of TREK-1 Channels". Cell Reports. 20 (6): 1287–1294. doi:10.1016/j.celrep.2017.07.034. PMC 5586213. PMID 28793254.
- Pavel, MA; Chung, HW; Petersen, EN; Hansen, SB (October 2019). "Polymodal Mechanism for TWIK-Related K+ Channel Inhibition by Local Anesthetic". Anesthesia and Analgesia. 129 (4): 973–982. doi:10.1213/ANE.0000000000004216. PMID 31124840.
- Hodgkin MN, Pettitt TR, Martin A, Michell RH, Pemberton AJ, Wakelam MJ (June 1998). "Diacylglycerols and phosphatidates: which molecular species are intracellular messengers?". Trends Biochem. Sci. 23 (6): 200–4. doi:10.1016/S0968-0004(98)01200-6. PMID 9644971.
- Wakelam MJ (December 1998). "Diacylglycerol--when is it an intracellular messenger?". Biochim. Biophys. Acta. 1436 (1–2): 117–26. doi:10.1016/S0005-2760(98)00123-4. PMID 9838074.
- Cockcroft S, Baldwin JM, Allan D (July 1984). "The Ca2+-activated polyphosphoinositide phosphodiesterase of human and rabbit neutrophil membranes". Biochem. J. 221 (2): 477–82. doi:10.1042/bj2210477. PMC 1144062. PMID 6089740.
- Moritz A, De Graan PN, Gispen WH, Wirtz KW (April 1992). "Phosphatidic acid is a specific activator of phosphatidylinositol-4-phosphate kinase". J. Biol. Chem. 267 (11): 7207–10. doi:10.1016/S0021-9258(18)42504-5. PMID 1313792.
- Bocckino SB, Blackmore PF, Wilson PB, Exton JH (November 1987). "Phosphatidate accumulation in hormone-treated hepatocytes via a phospholipase D mechanism". J. Biol. Chem. 262 (31): 15309–15. doi:10.1016/S0021-9258(18)48176-8. PMID 3117799.
- Hodgkin MN, Gardner SD, Rose S, Paterson A, Martin A, Wakelam MJ (March 1997). "Purification and characterization of sn-1-stearoyl-2-arachidonoylglycerol kinase from pig testes". Biochem. J. 322 (Pt 2): 529–34. doi:10.1042/bj3220529. PMC 1218222. PMID 9065773.
- Fleming IN, Yeaman SJ (June 1995). "Purification and characterization of N-ethylmaleimide-insensitive phosphatidic acid phosphohydrolase (PAP2) from rat liver". Biochem. J. 308 (Pt 3): 983–9. doi:10.1042/bj3080983. PMC 1136819. PMID 8948459.
- Olivera A, Rosenthal J, Spiegel S (March 1996). "Effect of acidic phospholipids on sphingosine kinase". J. Cell. Biochem. 60 (4): 529–37. doi:10.1002/(SICI)1097-4644(19960315)60:4<529::AID-JCB9>3.0.CO;2-U. PMID 8707892. S2CID 34752646.
- Morris AJ, Frohman MA, Engebrecht J (October 1997). "Measurement of phospholipase D activity". Anal. Biochem. 252 (1): 1–9. doi:10.1006/abio.1997.2299. PMID 9324933.
- Wiczer, Brian M; Thomas, George (27 Mar 2012). "Phospholipase D and mTORC1: Nutrients Are What Bring Them Together". Sci. Signal. 5 (217): pe13. doi:10.1126/scisignal.2003019. PMID 22457329. S2CID 206671479.
- Cabanos, C; Wang, M; Han, X; Hansen, SB (8 August 2017). "A Soluble Fluorescent Binding Assay Reveals PIP2 Antagonism of TREK-1 Channels". Cell Reports. 20 (6): 1287–1294. doi:10.1016/j.celrep.2017.07.034. PMC 5586213. PMID 28793254.
- Hite, RK; Butterwick, JA; MacKinnon, R (6 October 2014). "Phosphatidic acid modulation of Kv channel voltage sensor function". eLife. 3. doi:10.7554/eLife.04366. PMC 4212207. PMID 25285449.
- Hansen, SB; Tao, X; MacKinnon, R (28 August 2011). "Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2". Nature. 477 (7365): 495–8. Bibcode:2011Natur.477..495H. doi:10.1038/nature10370. PMC 3324908. PMID 21874019.